Fig. 5. HmK structure, interaction surface, and binding orientations.
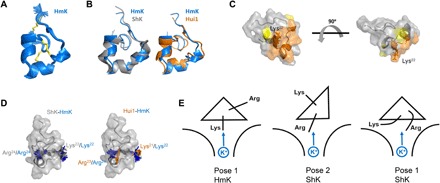
The HmK structure was determined from 328 distance and dihedral constraints derived from homo- and heteronuclear 2D NMR spectra (see Materials and Methods). (A) HmK structure (PDB accession code 6EI7). The 25 lowest-energy structures obtained from distance geometry/simulated annealing determination are shown. (B) Superposition of HmK (blue) with the crystal structure of ShK (4LFQ; gray) or the NMR structure of Hui1 (2N6B; orange) based on alignment of HmK/ShK residues 3 to 35 (Hui1, 2 to 34) shows that HmK retains an SAK1 scaffold. (C) Interaction surface of HmK mapped with T-toxin scanning as described in Fig. 3A. Orange indicates residues forming the primary toxin interaction surface (mutation caused ΔΔG > 2 kcal/mol). Yellow indicates Pro8 and Asn26 (mutation caused ΔΔG > 1.3 kcal/mol). The side chain of Lys22 is shown in dark orange. The two structures are separated by a rotation of 90o. (D) Left: Superposition of ShK and HmK highlighting ShK-Arg24 (gray)/HmK-Lys22 (blue) that mediate voltage dependence and ShK-Lys22 (gray)/HmK-Arg24 (blue) that do not. Right: Superposition of Hui1 and HmK highlighting Hui1-Arg23 (orange)/HmK-Lys22 (blue) that mediate voltage dependence and Hui1-Lys21 (orange)/HmK-Arg24 (blue) that do not. (E) Cartoon suggesting SAK1 toxin binding orientations in the hKv1.3 external vestibule with Lys or Arg near the conduction pore. Left: Pose 1 for HmK with Lys22 toward K+ in the pore. Middle: Pose 2 for ShK with Arg24 near K+ in the pore. Right: Pose 1 for ShK with Arg24 flexing to interact with K+ in the conduction pore.
