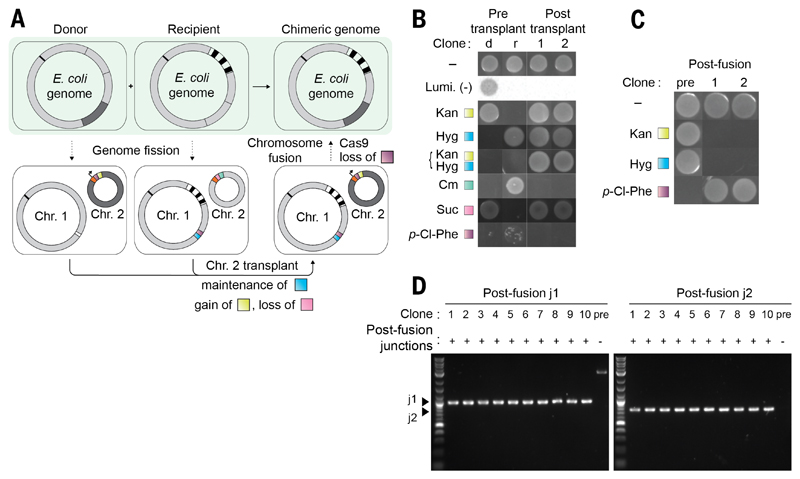
Fig. 4. Precise genome assembly from genomic segments of distinct strains.
(A) Precisely combining the watermarked region 1 (dark gray) from a donor strain and a watermarked region 2 (black striped) from a recipient strain into a single strain. Fission is performed in parallel in the donor and recipient strains. The resulting donor strain contains a watermarked Chr. 2 containing an oriT (black arrow) and a pheS*-KanR double selection cassette (purple and yellow); the remainder of Linker sequence 2 is orange. The resulting recipient strain contains an analogous non-watermarked Chr. 2, with a sacB-CmR cassette (pink and green). The linker sequence 1 (white) is replaced with a fusion sequence containing a pheS*-HygR cassette (purple and blue) in preparation for fusion. The donor cell is provided with a non-transferable F' plasmid. Mixing of donor and recipient cells facilitates conjugative transplant of Chr. 2 from the donor to the recipient; selection for KanR and against sacB-mediated sucrose sensitivity enables the isolation of cells that have gained a watermarked Chr. 2 and lost the non-watermarked Chr. 2. Subsequent genome fusion generates a strain in which the watermarked regions 1 and 2 have been precisely combined in a single chromosome. (B) Following chromosomal transplant by growth on selective media and luminescence. d, the pretransplant donor; r, pretransplant recipient. (C) After chromosomal fusion through growth on selective media. (D) PCR across the new junctions generated by chromosomal fusion yields products of the expected size in the post-fusion clones (1 to 10) but not in the prefusion control.