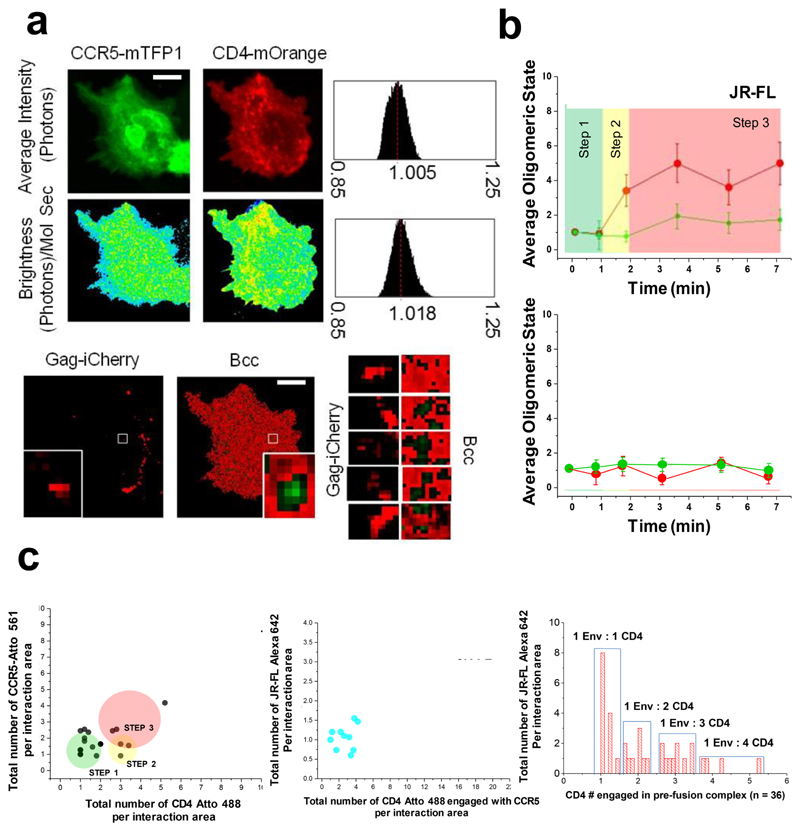
Figure 4. Visualization of JR-FL based HIV-1 virion receptor stoichiometry in live cells.
a, Fast time-resolved, three-color imaging was performed on a COS7 cell co-expressing CCR5-mTFP1 (green micrographs) and CD4-mOrange (red micrographs) exposed to HIVJR-FL –Gag-iCherry virions, scale bar 1 μm. The brightness histogram for each channel together with the pixel by pixel Brightnes maps are also presented (second row). In the last row, microsgraphs corresponding to the HIVHXB2-iCherry (in red) togher with the Bcc map is shown. Green regions represent positive Bcc. The time-resolved co-localization map coming from the white squares is also presented (third row, right panels) and correspond to regions of 0.5 X 0.5 μm. b, Time-resolved stoichiometry for CD4 (red dots) and CXCR4 (green dots) upon addition of HIVJR-FL/GagiCherry (right panel). The circles indicate the average value (n = 12) and the error bars indicate the standard error for each time point c, Time-resolved homotypic interactions for CD4 (red dots) and CCR5 (green dots) upon addition of HIVJR-FL/GagiCherry in the presence of inhibitory concentrations of b12. CD4 and CCR5 did not interact in this case. Eror bars indicate standard error for each time point which represents the average (n = 14). c, The total number of normalized events per interaction area of CCR5 labeled with Atto 561 was plotted against the total number of events per interaction area of CD4 labeled with Atto 488 for COS7 cells exposed to HIVJR-FL also labelled with Alexa 633 against the Env (as described in material and methods) (n = 23). The middle panel shows the total number of HXB2 Env interacting with CD4 and CXCR4 labelled with Alexa 405 are plotted against the total number of CD4 – CXCR4 interacting complexes (n = 12). The right panel shows the distribution of all CD4 – Env interactions and the relative frequency of their stoichiometry (n = 36).