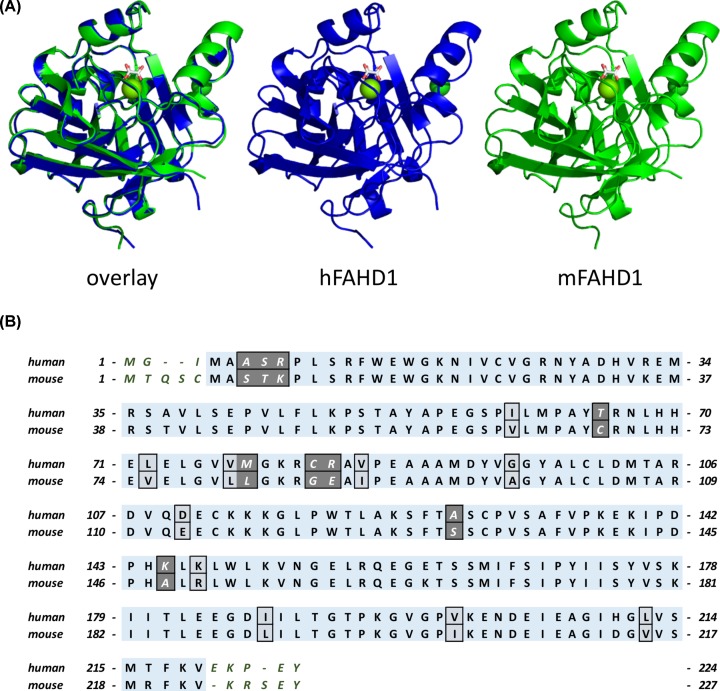
Figure 2. Overall structural comparison and sequence alignment of hFAHD1 and mFAHD1.
(A) Overlay and independent model structures of chain A in human (PDB: 6FOG) and mouse FAHD1 (PDB: 6SBI) after structural alignment. The general similarity is apparent, and further analysis (see Figure 3A), displays a low deviation among proteins in the unit cell. Sequence numbers in the aligned core proteins are offset by 3 due to the N-terminal differences. (B) Amino acid sequence alignment and comparison of human isoform 1 (UniProt: Q6P587) and mouse FAHD1 (UniProt: Q8R0F8). Boxes indicate differences in the amino acid sequences, as also listed in Table 3. Dark gray coloring thereby indicates changes in the chemical nature of the amino acid side chains.