Figure 1.
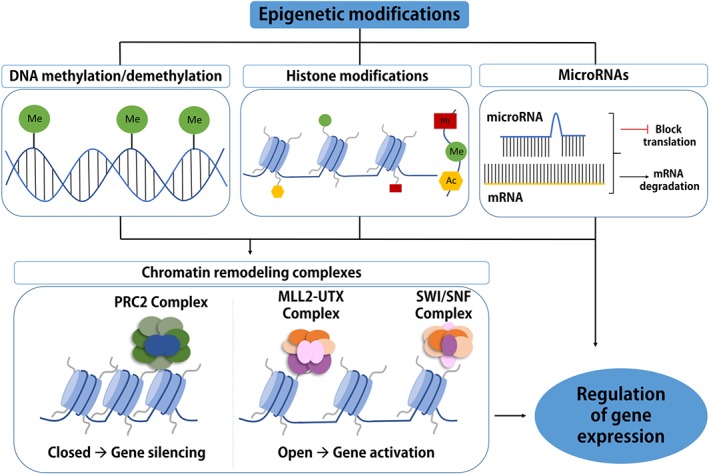
Epigenetic mechanisms of regulation of gene expression. Epigenetic regulation results in changes in gene expression without changes in the underlying DNA sequence. Epigenetic modifications include DNA methylation, histone modifications, non‐coding RNA‐related mechanisms, such as micro‐RNAs, and chromatin remodelling complexes. DNA methylation involves the covalent addition of a methyl group to the 5′ position of the cytosine base in DNA by DNA methyltransferases (DNMTs; DNMT1, DNMT3A and DNMT3B), and this reaction can be reversed by ten‐eleven translocation (TET) proteins and other proteins. Histone tails protruding from histone proteins can undergo acetylation (Ac), methylation (Me), phosphorylation (Ph), amongst other chemical modifications. Histone modifications are carried out by histone acetyltransferases (HATs) and histone deacetylases (HDACs), histone methyltransferases (HMTs) and histone demethylases (HDMs), Aurora B, and protein phosphatase 1 (PP1). miRNAs can epigenetically regulate gene expression at the post‐transcriptional level by binding to mRNA, which leads to targeted mRNA degradation and inhibition of gene expression. All these epigenetic mechanisms can influence chromatin remodelling (a condensed state or a transcriptionally accessible state), in addition to chromatin remodelling complexes such as polycomb repressive complex 2 (PRC2), MLL2‐UTX, and Swi/SNF. All of these mechanisms ultimately regulate gene expression
