Abstract
Mitochondria contribute to tumor growth through multiple metabolic pathways, regulation of extracellular pH, calcium signaling, and apoptosis. Using the Mitochondrial Nuclear Exchange (MNX) mouse models, which pair nuclear genomes with different mitochondrial genomes, we previously showed that mitochondrial single nucleotide polymorphisms (SNPs) regulate mammary carcinoma tumorigenicity and metastatic potential in genetic crosses. Here we tested the hypothesis that polymorphisms in stroma significantly affect tumorigenicity and experimental lung metastasis. Using syngeneic cancer cells (EO771 mammary carcinoma and B16-F10 melanoma cells) injected into wild-type and MNX mice [i.e., same nuclear DNA but different mitochondrial DNA (mtDNA], we showed mt-SNP-dependent increase (C3H/HeN) or decrease (C57BL/6J) in experimental metastasis. Superoxide scavenging reduced experimental metastasis. In addition, expression of lung nuclear-encoded genes changed specifically with mt-SNP. Thus, mitochondrial-nuclear cross-talk alters nuclear-encoded signaling pathways which mediate metastasis via both intrinsic and extrinsic mechanisms.
Keywords: Breast Cancer, Melanoma, Genetics, Latency, Metastasis, Metastasis efficiency, Mitochondria, Mitochondrial-Nuclear Crosstalk, Mitochondrial-nuclear exchange mice, Model, Polymorphism, Transgenic
Introduction
Otto Warburg’s discovery that cancer cells favor glycolysis followed by lactic acid fermentation over oxidative phosphorylation (1) has helped fuel research into mitochondria’s role in tumor formation and progression. It has since been shown that mitochondria contribute to tumor growth through multiple metabolic pathways, regulation of extracellular pH, calcium signaling, and apoptosis (2-6). Mitochondria have also been implicated in metastasis (reviewed in (7)). Several studies showed cybrid cells containing mitochondria from aggressively metastatic cell lines but nuclei from non-metastatic cell lines became more metastatic (8-10). These data, together with our previous findings (11), implicate mitochondria present within cancer cells, at least partially, determine cellular tumorigenicity and metastatic propensity.
Stephen Paget proposed that, in addition to the cancer cell itself which he termed the “seed”, the secondary microenvironment, the “soil”, also plays a role in determining whether cancer cells successfully metastasize to a tissue (12). Fidler and colleagues went on to show that, although vascular arrest plays a role in metastatic distribution as espoused by Virchow (13), without the proper microenvironment, metastatic cells fail to colonize (14). Coupled with our prior observations, we hypothesized that mitochondria contribute to the ability of the secondary site to support or undermine the growth of metastases.
Mitochondrial Nuclear Exchange (MNX) mice represent a unique opportunity to study host tissue/cell mitochondrial effects. MNX mice are made by enucleating a fertilized oocyte of one mouse strain, then transferring in a nucleus from a donor oocyte of a different mouse strain (15-17). For simplicity and clarity, MNX strain nomenclature is abbreviated (Table 1). By pairing known mouse metastatic cell lines with MNX strains that match the nuclear component of the cells but have different mitochondria (Table 2), mitochondrial host effects on metastasis can be studied.
Table 1:
Mouse strain nomenclature and abbreviations
| Strain | Nuclear DNA | Mitochondrial DNA | Abbreviation |
|---|---|---|---|
| FVB/NJ-mtMNX(FVB/NJ) | FVB/NJ | FVB/NJ | FF |
| FVB/NJ-mtMNX(C57BL/6J) | FVB/NJ | C57BL/6J | FC |
| FVB/NJ-mtMNX(BALB/cJ) | FVB/NJ | BALB/cJ | FB |
| BALB/cJ-mtMNX(BALB/cJ) | BALB/cJ | BALB/cJ | BB |
| C57BL/6J-mtMNX(C57BL/6J) | C57BL/6J | C57BL/6J | CC |
| C57BL/6J-mtMNX(C3H/HeN) | C57BL/6J | C3H/HeN | CH |
| C3H/HeN-mtMNX(C3H/HeN) | C3H/HeN | C3H/HeN | HH |
| C3H/HeN-mtMNX(C57BL/6J) | C3H/HeN | C57BL/6J | HC |
Mitochondrial nuclear exchange (MNX) mice were made by enucleating an oocyte of one mouse strain and transferring in a nucleus from a donor oocyte. Nuclear and mitochondrial contributors are listed next to their respective MNX strain. Abbreviations for each strain are defined.
Table 2:
Metastatic cell line origin, dosing, and incubation period
| Cell Line | Cancer Type |
Origin | Administered to |
Cell Number (Experimental) |
Cell Number (Spontaneous) |
Incubation Period (Experimental) |
|---|---|---|---|---|---|---|
| Pei1/MVT1 | Mammary | FVB/NJ | FF, FB, FC | 5 x 104 | 5 x 104 | 3 weeks |
| EO771 | Mammary | C57BL/6J | CC, CH | 5 x 104 | 1 x 105 | 2 weeks |
| B16-F10 | Melanoma | C57BL/6J | CC, CH | 1 x 105 | 1 x 105 | 2 weeks |
| K1735-M2 | Melanoma | C3H/HeN | HH, HC | 2 x 104 | 2 x 105 | 2 weeks |
Metastatic cell lines were paired with MNX mice which have matching nuclear backgrounds. These cells were injected into the tail vein (experimental) and ectopically (spontaneous) at the indicated dose. Time for metastatic formation used in experimental assays is listed.
Four different MNX strains and their wild-type counterparts were utilized in a series of metastasis assays. We show that mitochondria in non-cancer (i.e., stromal) compartments can affect metastasis in both mammary and melanoma models. Additionally, in vitro analyses showed variations in mitochondrial load, membrane potential, mitochondrial DNA copy number and metabolic profiles of mitochondria in the MNX and wild-type host tissue. We also showed that reactive oxygen species (ROS) scavenging of host tissue in vivo altered the metastatic propensity of these tissues. Finally, we used ex vivo approaches to analyze expression of metastatic genes with and without ROS scavenging to show that different mitochondrial backgrounds resulted in differential nuclear gene regulation. In turn, we showed that nuclear gene expression can be altered by mitochondrial ROS levels. Taken together these data show, for the first time, that mitochondria in the metastatic secondary environment affect the ability of metastatic cells to colonize. Although a definitive mechanism for this observation has yet to be solidified, our data indicated that ROS and mitochondrial-nuclear crosstalk play a role.
Materials and Methods
Mouse Lines
Stable MNX mouse lines were created as previously reported (17). Briefly, pronuclei were isolated from fertilized oocytes of FVB/NJ mice and transferred into enucleated fertilized oocytes of either C57BL/6J or BALB/cJ origin. Additional lines were created by exchanging pronuclei between oocytes of C57BL/6J origin and C3H/HeN origin. Wild-type FVB/NJ, C57BL/6J, BALB/cJ mice were purchased from Jackson labs, while C3H/HeN were purchased from Harlan laboratories (now Envigo). MNX colonies were maintained by breeding MNX females with nuclear matched male mice. All animal studies were approved by the Institutional Animal Care and Use Committee at the University of Kansas Medical Center (IACUC protocols 2014-2215; 2017-2408; 2017-2409)
Genotyping
Tail clips of no more than 3 mm in length were collected from all breeding and experimental mice at weaning. DNA extraction was performed using REDExtract-N-Amp Tissue PCR Kit (Sigma-Aldrich: XNAT-100). Restriction Fragment Length Polymorphism was used to ensure homoplasmic mitochondrial background as described (15, 16). Briefly, primers were designed to span regions of single nucleotide polymorphisms (listed below) which distinguish C57BL/6J (C9461T) mtDNA from FVB/NJ and C3H/HeN, and BALB/cJ (A9348G) mtDNA from FVB/NJ (18). The mutation at position 9461 in C57BL/6J mtDNA will not allow a restriction digest site for Bcl1 to incorporate. Upon incubation with Bcl1 (New England Biolabs: R0160S) cleavage will only occur in animals with FVB/NJ or C3H/HeN mitochondria. The mutation at position 9348 in BALB/cJ mtDNA results in the incorporation of a restriction digest site for Pflf1 (New England Biolabs: R0595S). Upon digest with Pflf1, cleavage will occur in animals with BALB/cJ mtDNA but not in those with FVB/NJ mtDNA.
In vivo Metastasis Assays
For experimental metastasis assays, syngeneic mouse metastatic cancer lines were injected into the tail vein of MNX and nuclear matched wild-type mice at four weeks of age (Table 2). Following an incubation of 3 (MVT1) or 2 (EO771, B16-F10, K1735-M2) weeks, mice were euthanized, and lungs were harvested upon necropsy. MVT1 cells were provided by Kent Hunter and Lalage Wakefield (NCI); K1735-M2 cells were gifted from Isaiah J. Fidler (M.D. Anderson Cancer Center); and, E0771 cells were provided by Linda Metheny-Barlow (Wake Forest Baptist Cancer Center). All cell lines were used within 10 passages following thawing; were validated by STR sequencing and were verified to be Mycoplasma free by PCR.
For spontaneous metastasis assays, syngeneic mouse metastatic cancer lines were ectopically injected (mammary fat pad for MVT1 and EO771 cells, intradermal for B16-F10 and K1735-M2) into 6-week old MNX and nuclear matched wild-type mice. Tumors were measured with digital calipers every other day and mice were euthanized when tumors reached mean diameter of 15 mm. Lungs were harvested upon necropsy.
Metastasis Quantification
For non-melanotic cell lines (MVT1, EO771, K1735-M2), lungs were stained for one hour in Bouin’s Solution (Sigma-Aldrich: HT10132) and rinsed twice in PBS. Melanotic B16-F10 colonized lungs were harvested then rinsed three times in PBS. Since 90% of lung metastases are visible on the surface (19), lungs were counted utilizing a dissecting scope and photographed. A non-parametric Kruskal-Wallis test followed by Dunn’s Method of pairwise multiple comparison was utilized to determine significance between MNX and wild-type groups.
Mitochondrial Load and Membrane Potential
To test mitochondrial load and membrane potential, mouse embryonic fibroblasts (MEF) were generated from all MNX and wild-type strains. MEF lines ( < passage 3) were harvested and stained individually and in combination with MitoTracker Red CMXRos (Molecular Probes by Invitrogen: M7512) and MitoTracker Green FM (Molecular Probes by Invitrogen: M7514) fluorescent probes followed by analysis by flow cytometry as described previously (20). Briefly, 100,000 cells from each line were added to cytometer tubes, and then stained with 200 nM of each probe alone or in combination. Cells from each line were also left as unstained controls. Cells were protected from light and the dye incubated for 15 minutes at 37o before placing on ice. Flow cytometry was performed with parameters from Molecular Probes website (MitoRed: Excitation-579 nm and Emission-599 nm, MitoGreen: Excitation-490 nm and Emission-516 nm). Fluorescence intensity at the appropriate emission wavelengths was captured.
Oxygen consumption and extracellular acidification
Mitochondrial flux was determined using the XF Seahorse Bioanalyzer and XF Cell Mitochondrial Stress Test Kit (Seahorse Bioscience: 101706) as described in the provided kit user guide. Briefly, MEF lines were cultured and passaged once prior to plating 40,000 cells per well in the provided Seahorse Plate. Cells were allowed to attach overnight. Sensor cartridge was loaded with Seahorse calibrant and placed at 37o without CO2 overnight. Cells were checked for confluent seeding then media was replaced with warmed unbuffered DMEM at pH 7.35. Cell plate was then incubated at 37o without CO2 for one hour. Measurements of oxygen consumption were taken before and after injections of oligomycin, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), antimycin A and rotenone (5 μM, 3 μM, 1.2 μM, and 1 μM, respectively). After Seahorse analysis was completed, cells were lysed and total protein concentration was analyzed using the Bradford method. Area under the curve for all measurements was normalized to total protein content.
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were also determined using the XF Seahorse Bioanalyzer with a subset of MNX MEF lines. The XF Glycolysis Stress Test Kit (Seahorse Bioscience: 103020) was utilized as described in the kit user guide. Briefly, MEF cells and sensor plate were loaded and incubated as listed above. Measurements of OCR and ECAR were taken before and after injections of glucose, oligomycin, 2-deoxy glucose (2-DG), antimycin A, and rotenone (10 mM, 1 μM, 100 mM, 2 μM, 1 μM, respectively). Cells were lysed and total protein concentration was used to normalize all oxygen consumption rate OCR and ECAR measurements.
Statistical significance was determined by first assessing equal variance through the Brown-Forsythe test followed by the Holm-Sidak pairwise multiple comparison test.
Total DNA Isolation
Total cellular DNA was isolated utilizing procedures that minimize loss of mitochondrial DNA as described previously (21). Briefly, equal weights of lung tissue were homogenized in a Tris/EDTA/SDS lysis buffer while on ice. Proteinase K (NEB: P81025S) was added to homogenate, samples were vortexed then incubated at 55o degrees Celsius for two hours. Samples were centrifuged at 16,000 x g for 15 minutes and supernatant was collected in new microcentrifuge tubes. Phenol:chloroform:isoamyl alcohol 25:24:1 solution (Sigma P3823) was added to supernatants and vortexed briefly. Samples were centrifuged and supernatant was collected in a new tube. Chloroform was added to supernatant, gently mixed by inversion (20x) then centrifuged. Supernatants were transferred into new tubes and sodium acetate and isopropanol were added. Samples were mixed gently by inversion (10x) and incubated at negative twenty Celsius overnight to allow DNA precipitation. Samples were centrifuged and supernatant was discarded. DNA pellets were washed twice with ethanol (70%) and air dried. DNA was suspended in nuclease-free water and DNA quantity and quality was analyzed via Nanodrop (ThermoFisher).
Mitochondrial DNA Content
To determine mitochondrial DNA copy number, lungs were harvested from untreated four-week old MNX and wild-type mice and snap frozen. Total cellular DNA (5 ng) was isolated and qPCR was performed with Taqman Fast Advanced MasterMix (Applied Biosystems: 4444557) and mouse specific Taqman probes. Expression levels of the nuclear encoded 18s ribosomal RNA (18s, ThermoFisher: Mm04277571), and mitochondrial-encoded genes NADH dehydrogenase subunit 1 (ND1, ThermoFisher: Mms04225274) and mouse cytochrome c oxidase subunit II (COX2, ThermoFisher: Mm03294838) were obtained using the Life Technologies ViiA 7 qPCR instrument. Fold-change for the mitochondrial genes was calculated and normalized to fold-change for nuclear 18s. To assess the statistical significance of differences in mitochondrial content between mouse strains a non-parametric Kruskal-Wallis Wallis test was performed followed by Dunn’s Method of pairwise multiple comparison was performed for both COX2 and ND1.
In vivo Metastasis Assays with Reactive Oxygen Species Scavenging
Four-week old MNX mice were randomized into two groups, control and treatment. This process was repeated with four-week old nuclear-matched wild-type mice. Treatment group animals were injected intra-peritoneally with 0.7 mg/kg of the mitochondrial specific superoxide scavenger MitoTEMPO (Enzo Life Sciences: ALX-430-150-M005) in 0.1% dimethyl sulfoxide (DMSO) 24 hr and again 1 hr prior to metastatic cell line injection. Control group animals received intra-peritoneal injections of 0.1% DMSO at 24 hr and 1 hr prior to metastatic cell line injection. Metastatic cells (K1735-M2 cells into HH and HC mice, EO771 cells into CC and CH mice) were introduced via the tail vein and allowed to incubate as described in Table 2. Mice were euthanized and lungs were harvested, stained, and metastases counted as above. Statistical significance of metastasis number between all groups was determined using a Kruskal-Wallis test followed by Dunn’s Method of pairwise multiple comparison.
Analysis of cell extravasation/seeding
K1735-M2 melanoma cells were detached using 2.5 μM EDTA in PBS. Cells (4 × 106) were then labeled with 5 μM CellTrace CFSE (Carboxyfluorescein succinimidyl ester; Invitrogen; C34554) in 2 ml of PBS for 10 min at room temperature. Cells were then washed with 10% media followed by PBS prior to being resuspended at a concentration of 2 × 105 cells/ml in HBSS. K1735-M2 cells (100 μl or 2 × 104 cells) were then injected i.v. into 4-week-old wild-type (HH) or MNX mice (HC). Lungs were harvested 24 hours post-injection with right lung lobes analyzed by flow cytometry and left lobes by immunofluorescence for CFSE positive cancer cells. Briefly, for flow cytometry analysis, 700,000 events were collected using the Acurri C6 cytometer with dead cells identified using propidium iodide and excluded from further analysis. For immunofluorescence, the left lung was fixed in 10% formalin, embedded and sectioned. Sections (3 × 20 μm sections approximately 100 μm apart) were analyzed from each mouse for detection of CFSE+ cancer cells. The number obtained from all three sections for each individual mouse were averaged and graphed in comparison to the uninjected controls.
Immunohistochemistry
Lungs from WT and MNX mice injected with cancer cells with or without the antioxidant mitoTEMPO were first fixed in Bouin’s solution for 20 min followed by overnight fixation in 10% formalin. Tissues were then maintained in 70% ethanol prior to embedding in paraffin. Tissue sections (5 μm) were stained using respective antibodies. Antigen retrieval was performed using EDTA Buffer (1 mM EDTA, 0.05% Tween 20, pH 8.0). Briefly, endogenous peroxidases were quenched using 3% H202 for 30 min. Tissues were blocked using 2.5% normal goat serum in phosphate buffered saline with 0.25% Tween 20 (PBST). Primary antibodies, CD4 (1:100; eBioscience; 4SM95), CD8 (1:100; eBioscience; 4SM15), or FoxP3 (1:100; eBioscience; FJK-16s) were incubated overnight at 4°C. Primary antibodies were detected using secondary goat anti-rat (Impress MP7404) according to manufacturer instructions. As a control, sections were stained with the biotinylated anti-rat antibody only. For CD4 and CD8 antibodies, images were acquired at 20x magnification and for FoxP3, images were acquired at 10x magnification. Percent DAB positivity was measured using the IHC Image Analysis Toolbox plugin for DAB isolation and quantification through the Image J analysis software (22). To determine relative metastatic area, FoxP3 Images were taken at 10x magnification and metastatic nodules were identified by dense hematoxylin staining. Non-metastatic regions were then masked using the positive and negative quick selection tool in photoshop and area was measured using image J.
Endogenous In-Vitro ROS Characterization
Characterization of endogenous ROS in mouse lung tissue was determined using the ratiometric mass spectrometry probe MitoB as described (23, 24). Briefly, CH, CC, HC, and HH mice (n=5) were divided into treatment and vehicle groups. Depending on group, mice were treated by intraperitoneal injection with either 0.7 mg/kg MitoTEMPO in 0.1% DMSO (as described above) or vehicle (0.1% DMSO in HBSS). After 24 hr mice were injected with a second dose of MitoTEMPO or vehicle and allowed to recover for 1 hr before intraperitoneal injection with 0.8 μM/kg MitoB (Cayman Chemical, Cat#17116). Six hours after MitoB injection, mice were euthanized, and lungs were harvested and flash frozen. A second cohort of untreated HH mice (n=24) were intraperitoneally injected with MitoB, then euthanized at 6 hr post-injection. Lungs harvested from untreated mice were used to generate a MitoB/MitoP (Cayman Chemical, Cat#17117) standard curve in duplicate described below.
Lungs (100 mg each) were combined with a solution of 60% (vol/vol) acetonitrile and 0.1% (vol/vol) formic acid in HPLC grade water and homogenized. Each sample was spiked with 10 μl of deuterated standards (d15-MitoB, Cayman Chemical, Cat#17470 and d15-MitoP, Cayman Chemical, Cat#19296). Standard curve lungs were additionally spiked with serially diluted MitoB (0, 1, 5, 10, 50, 100 μM) or MitoP (0.1, 0.5, 1, 2.5, 10, 25 μM). Samples were centrifuged for 10 min at 16,000 x g and supernatant was transferred to new tubes. Pellet was re-extracted, and supernatant was pooled with the previous sample. Samples were filtered using 0.22 μm PVDF filter, dried under vacuum, and stored at −80oC until ready for liquid chromatography tandem mass spectrometry (LC-MS/MS).
Samples were re-dissolved in a solution of 20% (vol/vol) acetonitrile, 0.1% (vol/vol) formic acid in HPLC grade water, vortexed then centrifuged for 10 min at 16,000 x g. Supernatant was transferred to sterile autosampler vials and MitoB:MitoP ratios were quantified by LC-MS/MS using an Ascentis Express column (C18, 5 cm × 2.1 mm, 2.7 μm, Sigma-Aldrich) for separation and a Waters Xevo TQ-S triple quadrupole mass spectrometer as mass analyzer. The LC parameters were as follows: autosampler temperature, 5°C; injection volume, 5 μl; column temperature, 50°C and flow rate, 400 μl/min. The LC solvents were solventA: 0.1% formic acid in water and solvent B: 95% acetonitrile + 0.1% formic acid. Elution from the column was performed over 10 min with the following gradient: t = 0, 5% solvent B; t = 2, 5% solvent B; t = 3, 60% solvent B; t = 7, 100% solvent B; t = 7.5, 100% solvent B; t = 8, 5% solvent B; t = 10, 95% solvent B. Mass spectra were acquired using positive-mode electrospray ionization operating in multiple reaction monitoring (MRM) mode. The capillary voltage was 3,000 V, and cone voltage was 50 V. Nitrogen was used as cone gas and desolvation gas, with flow rates of 150 and 600 l/h, respectively. The source temperature was 150°C, and desolvation temperature was 500°C. Argon was used as collision gas at a manifold pressure of 4.3 × 10−3 mbar. Precursor and product ion m/z, collision energies and source cone potentials were optimized for each transition using Waters QuanOptimize software. The MRM transitions were: MitoP: 370.2 > 107.1; d15-MitoP: 385.2 > 107.0; MitoB: 397.9 > 135.0; d15-MitoB: 412.9 > 135.1. The Kruskal-Wallis test followed by Dunn’s Method of pairwise multiple comparison was utilized to determine significance between MNX and wild-type treated and untreated groups.
Gene Expression Analysis of TEMPO Treated and Untreated Mouse Lung
Four-week old MNX mice were randomized into two groups, control (receiving 0.1% DMSO) and treatment (receiving 0.7mg/kg MitoTEMPO in 0.1% DMSO). This process was repeated with four-week old nuclear-matched wild-type mice. Groups received vehicle or MitoTEMPO intra-peritoneal injections, 24 hr later groups then received a second injection. One hour later mice were euthanized; lungs were harvested and then snap frozen. Lungs were homogenized using the Bead Bug Microtube Homogenizer. RNA was harvested using the RNeasy Microarray Tissue Kit (Qiagen: 73304) with Qiagen RNase-Free DNase set (Qiagen: 79254). cDNA was synthesized using the iScript cDNA Synthesis Kit (BioRad: 1708891). qPCR was performed in triplicate using the ViiA7 from Life Technologies, with ThermoFisher TaqMan Fast Advanced MasterMix and ThermoFisher mouse specific Taqman Primers for microRNA (miR) 199a (Mm04238139), miR125b (Mm04238249), fam120a (Mm01327068), dnmt1 (Mm01151063), ago2 (Mm03053414), sgtb (Mm00522889), scai (Mm00618853), rab6b (Mm00620652), and sod2 (Mm01313000).
Metabolomics
Lung and mammary tissue were isolated from both wild-type and MNX mice at six weeks of age and flash frozen in liquid nitrogen. Tissue (lung and mammary - 100 ug each; n=8) was shipped to Metabolon® for analysis. Briefly, the Discover HD4 Platform was employed to measure >700 metabolites and small biochemical species using liquid chromatography and gas chromatography/mass spectrometry . Pearson correlation values comparing selected metabolites were determined and more extensive analyses are still being completed.
Results
To determine whether metastasis is affected by host mitochondria, metastatic mammary and melanoma cells were introduced into MNX mice that had the same nuclear, but different mitochondrial, genomic backgrounds. As a control, tumor cells were injected into mice with both the same nuclear and mitochondrial genetic backgrounds as the cell line (i.e., wild-type). Cells were introduced either directly into the vasculature via the tail vein (experimental metastasis assay) or orthotopically (spontaneous metastasis assay). The spontaneous assay recapitulates the entire metastatic cascade; but, if tumor formation is affected, then metastasis colonization effects are hard to tease out. When paired with the experimental metastasis assay, which bypasses prior tumor formation steps, a more complete picture of the step(s) involved in metastasis affected by mitochondria is achieved. Each assay was repeated three times independently with a minimum of 10 mice per group. Data from representative experiments are shown.
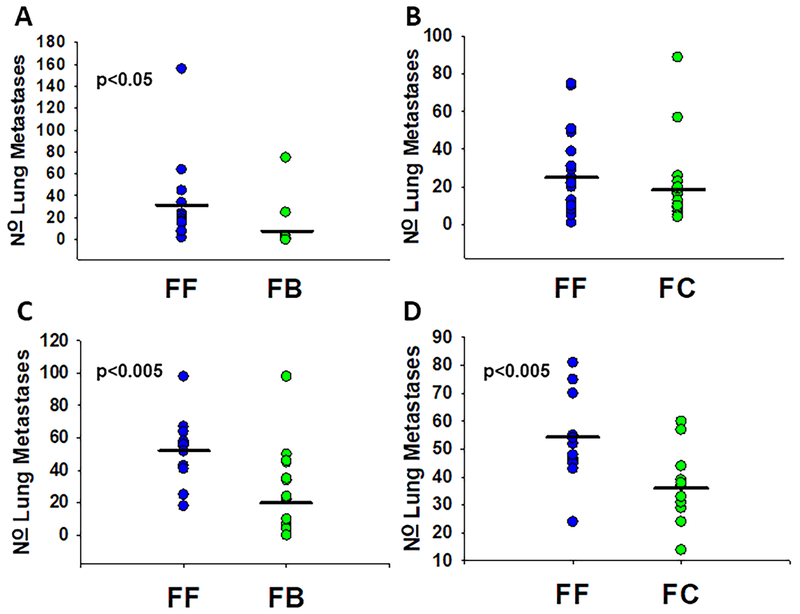
MVT1 metastatic mammary cells formed more lung metastases in control FVB/NJ animals than in MNX mice with BALB/cJ or C57BL/6J mitochondria. As seen in Figure 1A, FF mice (n=13) had a mean of 33 metastases while FB mice (n=11) had a mean of 11 metastases in the experimental assays. Following tail vein injections, the FF and FC groups did not show significant differences in the number of lung metastases (Figure 1B); however, the FF group had more metastases than the FC group, mean of 25 to 18 (n=20 and 21 mice, respectively). In spontaneous metastasis assays (Figure 1C), FF mice (n=14) had a mean of 52 metastases, while FB mice (n=20) had a mean of 20 metastases. MVT1 cells developed more lung metastases in the spontaneous assay (Figure 1D) in wild-type FF mice than in FC mice ( = 55 and 36; n=13 and 14, respectively).
Figure 1:
Mitochondrial haplotype alters total pulmonary metastases of mammary cells in spontaneous (orthotopic) and experimental (intravenous) models. Pei1 metastatic mouse mammary cells were injected into the A & B: tail vein and mice were euthanized three weeks after injection. C & D: Pei1 cells were also injected into the mammary fat pad and lungs were harvested once primary tumors reached 15 mm in average diameter. Upon euthanasia lungs were harvested and gross metastases were counted. Individual dots represent total number of metastases for each individual animal, black bars represent mean for the mitochondrial group. A: Tail vein injected FF mice (n=13 mice) had more ( = 33, SEM 11) metastases than FB mice (n=11, = 9, SEM 7). B: FF and FC tail vein injected mice did not differ in their total number of metastases (n=20, mean 25, SEM 5 and n=21, mean 18, SEM 4 respectively). C: Orthotopically injected FF mice (n=14) had more metastases ( = 52, SEM 5) than similarly injected FB mice (n= 20, = 20, SEM 6). D: Orthotopically injected FF mice (n=13) had more metastases ( = 55, SEM 4) than similarly injected FC mice (n=14, = 36, SEM 3).
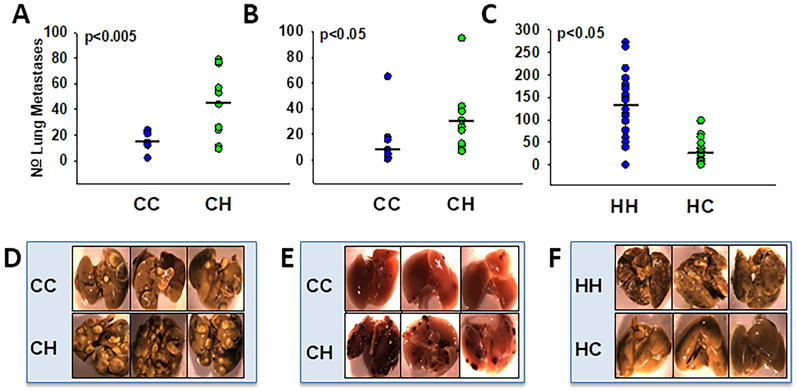
Mice with C3H/HeN mitochondria developed more metastases than mice with C57BL/6J mitochondria when utilizing either mammary or melanoma cancer lines. In Figure 2A, the mouse metastatic mammary line, EO771, was introduced via the tail vein into both wild-type C57BL/6J mice as well as CH MNX mice. CC mice formed on average 15 lung metastases while CH mice had an average of 45 lung metastases. When the B16-F10 melanoma cells were introduced to the same mouse strains, CC mice again had fewer metastases ( = 12) than CH mice ( = 29) (Figure 2B). In parallel experiments switching nuclear background to C3H/HeN, more metastases were still observed when mitochondria were C3H/HeN rather than C57BL/6J when utilizing K1735-M2 melanoma cells (Figure 2C). Figure 2D-F present representative data from Figures 2A-C.
Figure 2:
Mitochondrial haplotype alters metastatic propensity in both mammary and melanoma models. Metastatic mouse cell lines were injected into the tail vein of nuclear matched MNX mice. Tail vein injected mice were euthanized two weeks after injection, lungs were isolated and photographed top and bottom (representative photos shown in D, E, F) prior to metastasis quantification. Circles represent total number of metastases in each animal; black bars represent average number of metastases for the mitochondrial group. A, D: EO771 metastatic mammary cells injected into the tail vein of CC mice (n=9) resulted in fewer ( = 15, SEM 2) metastases than CH mice (n=10, = 45, SEM 9). B, E: B16-F10 metastatic melanoma cells injected into the tail vein of CC mice (n=10) showed fewer ( = 12, SEM 6) metastases than CH mice (n=10, = 29, SEM 8). C, F: K1735-M2 cells injected in the tail vein of HH mice (n=20) resulted in more metastases ( = 134, SEM 16) than similarly injected HC mice (n=20, = 31, SEM 7).
The cell lines, EO771, B16-F10, and K1735-M2 were also evaluated in spontaneous metastasis assays. EO771 were introduced into the mammary fat pad while B16-F10 and K1735-M2 were injected intradermally. All cell lines rapidly formed large necrotic tumors. Due to the fast tumor growth, most mice failed to develop overt, macroscopic metastases before they had to be euthanized for ethical reasons.
To begin exploring whether differences in mitochondrial mass or baseline metabolic changes in stroma contributed to changes of metastatic susceptibility in MNX mice, MEF were isolated and characterized for membrane potential, mitochondrial load (i.e., mitochondria number), OCR and ECAR, and mtDNA copy number.
The number of mitochondria present as well as the membrane potential did not differ significantly between MNX and wild-type animals analyzed. Using the fluorescent probes, MitoTracker Red, whose accumulation in mitochondria is dependent upon membrane potential, and MitoTracker Green, which accumulates in mitochondria independent of membrane potential, flow cytometry was performed with MEF cell lines created from each MNX and wild-type strains. As seen in Figure S1A, there was no statistically significant difference in overall number of mitochondria or membrane potentials between the different wild-type and MNX strains. Even when membrane potential was normalized to the number of mitochondria (as is routine for this type of measurement), there was no statistically significant difference between strains (Figure S1B).
OCR of MNX and wild-type MEF lines showed differences between the strains using the Seahorse Bioanalyzer. MEF lines made from a single embryo per line were analyzed for oxygen consumption in quadruplicate (Figure S2A) or quintuplicate (Figure S2B). As seen in Figure S3A, FC and FF cells had higher basal respiration = 95.725, 95.630 pmol/min/μg respectively) and maximal respiration ( = 175.604, 152.906 pmol/min/μg, respectively) than BB (basal respiration = 61.636 pmol/min/μg, maximal respiration = 81.228 pmol/min/μg) cells. ATP-Linked oxygen consumption rate did not appear to differ significantly among strains. No significant differences were seen between oxygen utilization profiles of CC, CH, HH, and HC mice (Figure S2B). Although differences in metabolic flux occurred between the strains, these differences did not appear to mirror the metastatic phenotypes shown above.
OCR and ECAR profiles of MEF lines isolated from MNX and wild-type mice showed differences between strains. Single embryo-derived MEF lines from CC, CH, HH, and HC mice were analyzed for ECAR in quintuplicate using the Seahorse instrument (Figure S3). Non-glycolytic acidification rates differed with CH having the highest ( = 1.437 mpH/min/μg) and HH having the lowest ( = 1.178 mpH/min/μg). Reserve capacity also differed between CC-derived cells ( = 1.089 mpH/min/μg) and HC-derived cells ( = 2.663 mpH/min/μg). Although slight variations were present, taken as a whole, OCR and ECAR between these four strains were remarkably similar. The differences observed did not parallel metastatic phenotypes.
mtDNA copy number was evaluated to determine if differences exist when dissimilar nuclear and mitochondrial genomes were combined. DNA from lung samples were isolated and expression of two mitochondrial specific genes (mtCO2, and ND1) were analyzed and normalized to expression of a nuclear specific gene (18s). As shown in Figure S4, HH and HC strains had a slightly higher (30%) mitochondrial copy number than CC or CH strains. Unfortunately, variance was too high to observe a statistically significant difference.
Mitochondrial ROS have been implicated in metastatic disease (25, 8, 9, 26-28). To determine if ROS were playing a role in metastatic susceptibility seen with varying mitochondrial genomes, we utilized a mitochondrial superoxide scavenger, MitoTEMPO. MitoTEMPO acts as a superoxide dismutase 2 mimic and specifically scavenges superoxides produced in the mitochondria (29, 30) when complex I and III reduce oxygen. Superoxide dismutates to longer lived hydrogen peroxide, which can partially reduce to hydroxyl radicals, or can react with nitrogen to form nitric oxide or peroxynitrite species (31). By targeting superoxide, we targeted the most upstream oxidant.
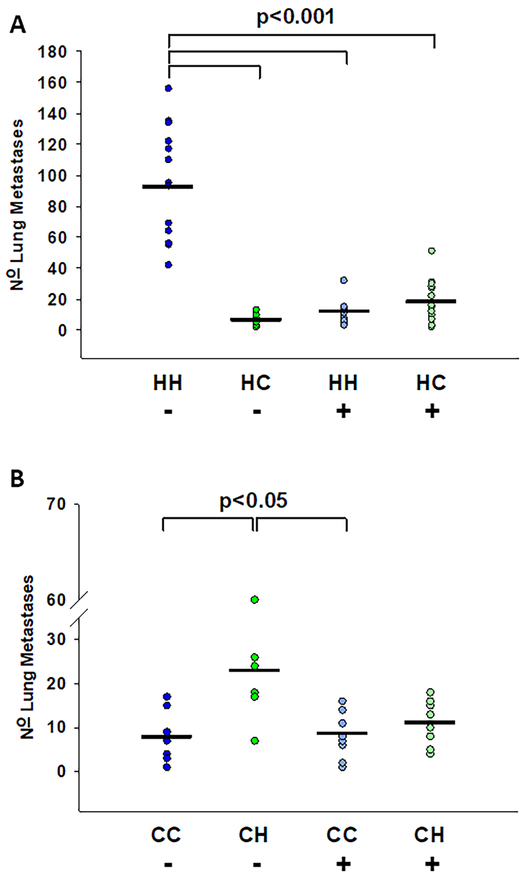
When MNX and wild-type mice were pre-treated with MitoTEMPO or vehicle twenty-four and again 1-hour prior to intravenous injection of tumor cells, metastasis was suppressed in all mice with C3H/HeN mitochondria. When K1735-M2 melanoma cells were used, numbers of metastases in the vehicle-treated mice matched those in untreated animals. HH mice developed an average of 94 metastases while HC developed only 16 metastases (Figure 3A). When treated with MitoTEMPO, HH mice developed a mean of 11 metastases while HC mice had 18 lung colonies. As shown in Figure 3B, when EO771 was injected after superoxide scavenging in the host, CH mice had reduced metastases ( = 11) compared to vehicle treated CH mice ( = 24), but the differences were not statistically significant. Regardless of cellular genetic background, MitoTEMPO reduced lung colonization to a level similar to that observed in mice with C57BL/6J mtDNA. MitoTEMPO treatment did not eliminate metastases but did reduce both the size and number of metastases (Figure S5). C3H/HeN mitochondria therefore appear to produce higher endogenous levels of ROS that, in turn, affect the seeding of circulating tumor cells in lungs.
Figure 3:
Mitochondrial superoxide scavenging selectively decreases metastasis in C3H/HeN mitochondrial mice. DMSO vehicle (−) or MitoTEMPO (+) was IP injected into A: 4-week old HH (− n=14 (vehicle only), + n=15 (MitoTEMPO)), and HC (− n=10, + n=15) mice 24 hours and again 1 hour prior to IV injection of K1735-M2 cells and B: 4 week old CC (− n= 9, + n=10), and CH (− n=8, + n=9) mice 24 hours and again 1 hour prior to IV injection of EO771 cells. Mice were euthanized two weeks post cell injection, lungs were harvested, and gross pulmonary metastases were quantified. A: K1735 injected HH vehicle mice had significantly more metastases ( = 94, SEM 10) than HC vehicle ( = 16, SEM 3), HH MitoTEMPO ( = 11, SEM 2), and HC MitoTEMPO ( = 18, SEM 4) treated mice. B: EO771 injected CH vehicle treated mice had more metastases ( = 24), than CC vehicle ( = 8), CC MitoTEMPO ( = 9), and CH MitoTEMPO ( = 11) treated mice.
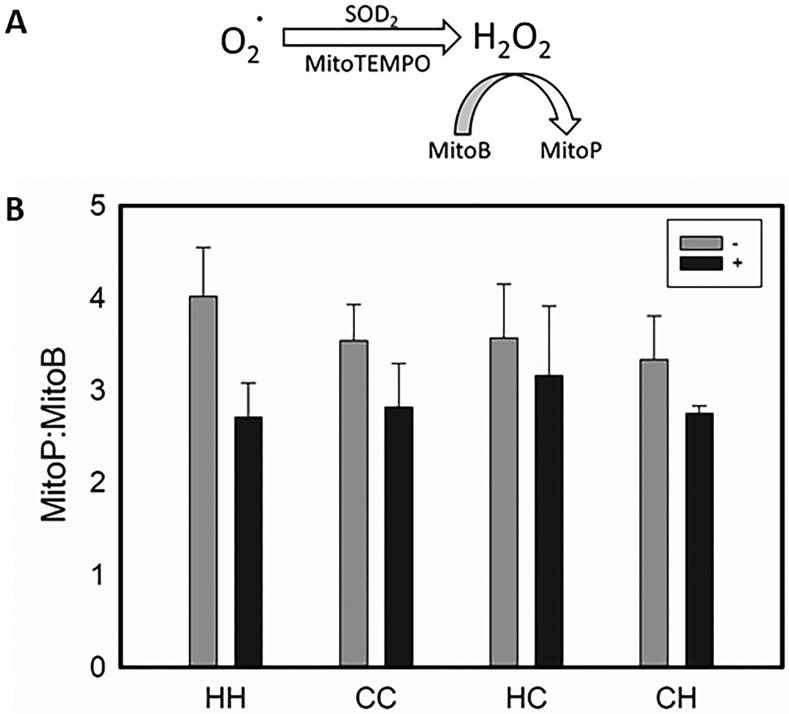
To determine if single nucleotide polymorphisms (SNP) in C3H/HeN mitochondria indeed produced higher basal levels of ROS than C57BL/6J, endogenous ROS was measured in lungs using MitoP:MitoB ratio (23, 24). Importantly, the reaction is stable after euthanasia and tissue extraction (Figure 4A). ROS varied between wild-type and MNX mice, but the differences were not statistically significant. MitoTEMPO treatment, however, always lowered ROS in lungs (Figure 4B). So, while C3H/HeN mitochondria appear consistently to make more ROS when coupled with C3H/HeN nuclear DNA, the difference is attenuated when C57BL/6 and C3H/HeN genomes are mixed. Measurement of ROS at specific locations (i.e., a pre-metastatic niche) could have identified foci of high ROS, but the ability to assess the tissues in this way are not sufficiently robust.
Figure 4:
Endogenous levels of reactive oxygen species vary with mitochondrial and nuclear background. A: MitoTEMPO acts as a SOD2 mimic to dismutate superoxide to hydrogen peroxide within the inner membrane of the mitochondria. The mitochondrial specific ratiometric mass spectrometry probe MitoB reacts with hydrogen peroxide to form the stable phenol, MitoP. B: DMSO vehicle (−) or MitoTEMPO (+) was IP injected into 4-week old HH (− n=5 (vehicle only), + n=5 (MitoTEMPO)), CC (− n=5, + n=5), HC (− n=5, + n=5), and CH (− n=5, + n=5) mice 24 hours and again 1 hour prior to IV injection of MitoB. Mice were euthanized 6 hours post MitoB injection, lungs were harvested, and the ratio of MitoP to MitoB was quantified using LC-MS/MS. Error bars represent standard error of the mean.
To assess whether ROS could alter nuclear gene expression, we first determined if expression of select metastasis-associated nuclear genes corresponded with mitochondrial haplotype. qPCR was performed using RNA isolated from lung tissue collected from CC, CH, HH, and HC strains to measure expression of miR199a, miR125b, fam120a, dnmt1, ago2, sgtb, scai, rab6b and sod2 (Figure S6). These genes were chosen because they exhibited significant differential expression and nuclear DNA methylation patterns in previous experiments (32). Most differences in basal expression varied by less than 2-fold. Interestingly, expression of all genes, except rab6b, was lower in CC compared to HH mice. All of these genes were more highly expressed in CH than CC mice. sod2 expression did not change significantly between CC, HH, CH or HC mice. Following MitoTEMPO treatment, expression of the same genes was measured to assess whether ROS might be mediating the effects. MitoTEMPO treatment raised expression levels for miR199a, miR125b, dnmt1, ago2, fam120a, rab6b, sgtb, and scai for both CC and CH mice (Figure S7A-I). Expression in HH and HC mice was unaffected by MitoTEMPO.
Increasing vascular permeability is important for immune extravasation into the lungs, which is further driven by increasing levels of ROS released by neutrophils or other immune cells (33). To begin addressing roles for vascular permeability, K1735-M2 cells were labeled with CFSE and introduced intravenously into HH or HC mice. After 24 hr, lungs were analyzed for CFSE positivity compared to uninjected controls using both flow cytometry and immunofluorescence. No significant differences were observed between the HH or HC mice (Figure S8), suggesting that differences in metastatic efficiency are likely a result of mtDNA differences existing within cells of the lung microenvironment rather than differences associated with trapping or early retention.
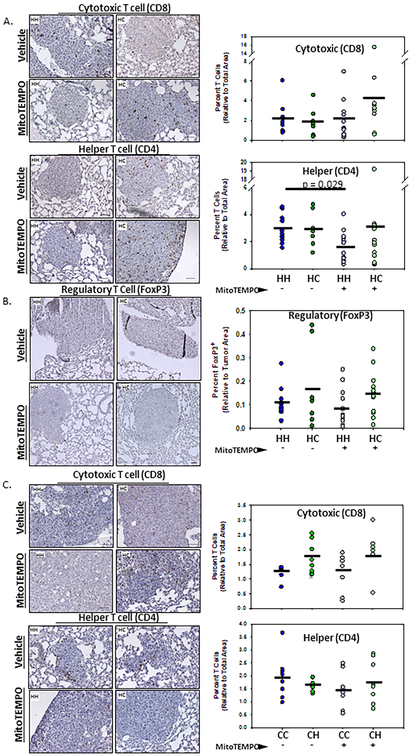
To begin addressing a potential role for mtDNA in regulating immune surveillance of the metastatic cascade, tissue sections were analyzed for differences in cells representing adaptive immunity. E0771 lung metastases exhibited a lower percentage of infiltrated CD8+ T cells in the WT (CC) (1.2%) vs MNX (CH) (1.7%) mice (P = 0.07). Interestingly, the percentage of CD8 cells were not altered by the treatment of WT (CC) (1.3%) and MNX (CH) (1.8%) mice with MitoTEMPO (Fig. 5 A). E0771 metastases had a higher percentage of CD4+ T cells in the WT (CC) (2%) vs the MNX (CH) (1.6%) background. Interestingly, MitoTEMPO reduced the percentage of T cells in the WT (CC) (1.5%) and increased the percentage of T cells in the MNX (CH) (1.8%) background (Fig. 5 A). Similar analyses were done for K1735-M2 metastases. No differences in T cell percentages between the WT (HH) (1.9%; 3%) and MNX (HC) (1.7%; 2.9%) backgrounds were observed for CD8+ and CD4+ T cells, respectively. MitoTEMPO did not alter CD8+ T cells (2%) in WT (HH) mice; however, CD8+ T cells did increase (to 4.2%) when MNX (HC) mice were treated with MitoTEMPO (Fig. 5 B). MitoTEMPO decreased the percentage of CD4+ T cells in the WT (HH) (1.6%) but not in the MNX (HC) (3.1%) mice (Fig. 5B). HH and HC lung sections were examined for the T regulatory cells using the transcription factor FoxP3. As above, Treg cells trended toward fewer in HH mice treated with MitoTEMPO [Vehicle = 1.05% vs. MitoTEMPO = 0.081%] (Fig. 5C).
Figure 5. Mitochondria stromal haplotype alters immune cell recruitment to sites of metastases..
Immunohistochemistry (IHC) analysis of formalin-fixed paraffin-embedded experimental lung metastases derived from A. K1735-M2 melanoma or C. E0771 mammary cancer cells for CD8+ cytotoxic T cells and CD4+ helper T cells. DMSO vehicle (−) or MitoTEMPO (+) was IP injected into 4-week-old mice 24 hours and again 1 hour prior to IV injection of cancer cells. Mice were euthanized two weeks post cell injection, lungs were harvested, and gross pulmonary metastases were quantified. A & C. Images obtained at 20x magnification. Quantification of percent CD8 and CD4 T cell positivity using IHC Image Analysis Toolbox plugin for DAB isolation and quantification through the Image J analysis software [PMID: 27121383]. Data as means (n = 6-14; One-way ANOVA and multiple comparisons T-test). B. Images obtained at 10x magnification. Quantification of percent FoxP3 cell positivity using IHC Image Analysis Toolbox plugin for DAB isolation and quantification through the Image J analysis software [PMID: 27121383]. Data as means ± SEM (n = 9-16).
The antioxidant glutathione protects cells from damage caused by ROS. In order to begin assessing the effects of ROS on metabolism, a metabolomic analysis was performed on six-week old lung and mammary tissue collected from FF, FC, FB, CC, CH, HH, HC. Both oxidized and reduced forms of glutathione showed minimal changes between wild-type and MNX mice. The only significant change observed was reduced glutathione (GSH), which was higher in CH compared to HH mice. There were significant changes observed when measuring the amino acids that feed into the gamma-glutamyl pathway between CC, CH, HH and HC. In general, however, none of these changes consistently correlated with altered metastatic propensity.
Discussion
Having previously demonstrated that nuclear (34) and mitochondrial (16, 11) genetics could alter the efficiency of mammary tumor development and metastasis, we sought to distinguish the impact of stromal contributions in metastatic colonization. Accumulating data implicate mitochondrial function (9, 35, 27, 36, 37) and localization (38) in the metastatic process. Since metastasis involves a complex interplay between tumor cells and stroma, including bi-directional signaling and recognition between cells, matrices and soluble factors, our experimental design kept nuclear and mitochondrial genomes constant between tumor cells and mice, while changing the mitochondria only in the stroma.
Using four different experimental models representing two histotypes (two mammary carcinomas, two melanomas), orthotopic tumor growth was not dramatically affected by mitochondrial background. In contrast, colonization of lung following intravenous inoculation of tumor cells was significantly affected by stromal mitochondria. C57BL6/J mitochondria always diminished metastasis, while C3H/HeN mitochondria consistently increased lung colonization. These results highlighted that both intrinsic and extrinsic mitochondrial genetic changes are important in tumor cell behavior.
The mouse strains used in this report have mtDNA that is quite similar. There are only five SNP distinguishing FVB, BALB/c, C57 and C3H mitochondria (18). Most SNP are in subunits of electron transport chain genes. FVB mitochondria have a G7777T SNP in ATPase subunit 8 which changes the aspartic acid residue to a tyrosine. FVB and C57 mitochondria have a A9348G transition in cytochrome oxidase subunit 3, which is the main transmembrane subunit of Cytochrome C Oxidase (complex IV). Despite this mutation coding for valine instead of isoleucine, it is thought to be a neutral mutation (18, 39). C57 have a C to T transversion at 9461 located at the start site for NADH Dehydrogenase subunit 3 (part of Complex I) which is non-synonymous. Interestingly, C57 and C3H have mutations in a mitochondrial tRNA (mt-tRNAArg). C57 mitochondria have an adenine deletion at position 9821 in mt-tRNAArg, while C3H mitochondria have an additional thymine located at 9820.
Mutations in electron transport subunits implicate metabolism. Previously, oxygen consumption was found to be different in MNX strains using mammary tissue (16) and cardiomyocytes (15), which potentially explained differences in the phenotypes observed. However, in the current experiments, we measured membrane potential, overall mitochondrial load, OCR and ECAR in MEF cell lines and did not find significant differences for any of these parameters that corresponded with changed tumor cell behavior. These results highlight a critical caveat regarding interpretation of these (or any) metabolism or gene expression results – metabolism, transcriptomes and mitochondrial number vary greatly from tissue to tissue (40-42). To properly assess stromal metabolic contributions to changing metastatic efficiency, full metabolomic profiles in every tissue encountered throughout the metastatic cascade would be required. Preliminary results comparing mammary and pulmonary tissue show remarkable similarities with some differences. However, those results require replication and validation.
While not statistically significant, we observed almost 30% more mtDNA in strains with the nuclear background originating from the C3H mouse, regardless of mitochondrial origin. Nuclear factors clearly influence the amount of mitochondrial coded DNA, but quantity of mtDNA in stromal cells did not fully explain changes in lung colonization.
While reconstructing the mutational history of the L929 leukemia cell lines, the Wallace laboratory found that mitochondrial mutations increased ROS production and proliferation (43). They postulated that this might be a mechanism by which mitochondria contribute to neoplasia. Using cybrids, others went on to show that additional adenines in the DHU loop of mt-tRNAArg increase ROS production and, in turn, proliferation and invasion (39, 44). Those observations compelled the hypothesis that altered ROS in MNX mice might be responsible for the changes in metastatic potential, especially since C3H has the most adenines in the DHU loop, while C57 have the fewest.
To test whether ROS differences could explain metastasis differences, we treated mice with MitoTEMPO, a scavenger for mitochondrially derived ROS. Treatment of mice prior to tumor cell injection dramatically reduced lung colonization in C3H mice but had a lesser effect in C57 mice. These results were consistent with the prediction that C3H mice would have higher baseline levels of ROS. However, complementary experiments to increase ROS in C57 mice would have too many deleterious effects on other physiologic systems (e.g., tissue damage) to safely and unequivocally interpret any findings.
To corroborate the interpretation regarding ROS levels, we utilized the recently developed MitoP:MitoB assay (23, 24). ROS were higher in HH mice, but not in CH compared to CC mice. Two interpretations are consistent with these observations. First, there may be a critical threshold level of ROS that would change metastatic efficiency. Second, ROS levels are also controlled by proteins in the mitochondria that are nuclear encoded. Additionally, the nucleus encodes other ROS-modifying machinery. Importantly, MitoP does not measure all ROS species, only H2O2 production and accumulation. Also, it is possible that there are non-ROS, off-target actions of MitoTEMPO (which have yet to be defined) that influence the stromal tumor microenvironment of a metastatic niche.
Accumulating evidence implicates ROS in several biochemical and physiologic processes important to metastasis. ROS have recently been shown to act as secondary messengers to the nuclear genome (45-59) where they alter gene expression, cell proliferation and growth. Data in this and our previous report (32) showed that not only can gene expression differ when mitochondrial background is changed, but nuclear-mitochondrial combinations result in unique gene expression profiles (32). Active mitochondrial-nuclear crosstalk thus defines nuclear gene expression. To determine if ROS could be a messenger for this cross-talk, we also evaluated expression profiles of mice treated with a ROS scavenger. We found that changing ROS levels alter expression of specific genes from specific backgrounds (Figure S7), but the scope of genes examined does not yet fully explain changes in metastatic potential.
Since MNX mice were generated by the transfer of mitochondrial DNA and not prolonged co-evolution of nDNA and mtDNA, some have questioned whether the findings are artifactual in nature. We believe that this is not the case for several reasons. First, male MNX mice do not transfer the mitochondrial genetic information to progeny (11). Second, the phenotypes observed have been highly stable over multiple generations of MNX mice. Third, early data using a subset of congenic backcrossed mice have yielded equivalent results to those reported here.
Complex genetic phenotypes, like metastasis, require coordinated expression of multiple genes (60-62). Moreover, the process of metastasis requires transmission of signals from external sources, like tissues, blood, and extracellular matrices. This concept was first articulated by Stephen Paget in the “Seed and Soil” hypothesis to explain organotropism of cancer metastasis (63). He concluded that metastasis is influenced by both intrinsic (cell autonomous) and extrinsic (non-cell autonomous) factors. The findings reported here establish, for the first time to our knowledge, that mitochondrial genetics of stromal cells influence metastasis from mammary carcinomas and melanomas to the lung. Early indications are that ROS from the stroma could be, at least partially, responsible for efficiency of lung colonization. And since ROS is integral to immune functions and activity, some studies to explore whether those connections exist within the MNX mice were performed. Although much more study will be required, analysis of post-mortem lung sections has provided preliminary evidence for differences in the immune landscape. We chose to focus on the adaptive immune system as the tissues were collected two weeks post- injection. Whereas our data suggests CD4+ T regulatory cells may play a role in ROS mediated differences in lung metastatic susceptibility, studies to more effectively determine the complex immune networks responsible for these phenotypes are ongoing. Although this study focused on ROS-related mechanisms to explain the mitochondrial SNP-associated changes in metastasis, we acknowledge that there remain alternative explanations which need to be more fully explored. The differences in immune profiles, for example, could also be affected by metabolites that alter immune cell polarization and/or immune cell cross-talk. Likewise, ROS are established mediators of endothelial patency and vascular permeability. Trans-endothelial migration efficiency or effects on other components of the reticuloendothelial system could also be involved.
Nonetheless, taken together, these findings have important implications in oncology. Specifically, our results imply that mitochondrial polymorphisms could, perhaps, be used to predict development of metastasis. And, since mitochondrial SNP can be used to define race in humans or strains in mice (64), they may partly explain racial disparities in cancer incidence and aggressiveness. Future experiments will explore these possibilities.
Supplementary Material
Significance Statement:
Stromal mitochondrial polymorphisms affect metastatic colonization through reactive oxygen species and mitochondrial-nuclear crosstalk.
Acknowledgments & Funding Support
The studies reported here were completed in partial fulfilment for a PhD degree in Molecular Physiology (AEB). The authors appreciate the superb technical support from Adam Scheid and Jonas Rowland (flow cytometry), Justin King (ImageJ analysis) and Thuc Ly (injections) as well as their helpful comments and suggestions. The authors also wish to thank Russell Swerdlow and Heather Wilkins for assistance with studies involving the Seahorse Bioanalyzer; Kent Hunter and Lalage Wakefield for providing the MVT1 cells; Isaiah J. Fidler for the K1735 cells; Linda Metheny-Barlow for the E0771 cells and Michael P. Murphy for helpful advice regarding the MitoB experiment. The authors acknowledge the superb technical support and advice from the Biostatistical and Bioinformatics Shared Resource of the University of Kansas Cancer Center.
Research supported by the following: Susan G. Komen for the Cure (SAC11037) (D.R. Welch); National Foundation for Cancer Research (D.R. Welch); NIH grants R01-CA134981 (D.R. Welch), P30-CA168524 (D.R. Welch, D.C. Koestler); Kansas Bioscience Authority (D.R. Welch); Biomedical Research Training Program Fellowship (A.E. Brinker); 2016 AACR-Incyte Corporation NextGen Grant for Transformative Cancer Research, Grant Number 16-20-46-LUNT (S.Y. Lunt); Office of the Assistant Secretary of Defense for Health Affairs, Breast Cancer Research Program, W81XWH-15-1-0453 (S.Y. Lunt) and W81XWH-18-1-0450 (T.C. Beadnell).
Footnotes
Conflict of Interest Disclosure Statement
D.R. Welch is a co-holder of a patent on the MNX mice but has received no financial support. The other authors declare no financial conflicts of interest
References:
- 1.Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. J Gen Physiol. 1927;8:519–30. Epub 1927/03/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang C, Youle RJ. The role of mitochondria in apoptosis*. Annual review of genetics. 2009;43:95–118. Epub 2009/08/08. doi: 10.1146/annurev-genet-102108-134850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinberg F, Chandel NS. Mitochondrial metabolism and cancer. Annals of the New York Academy of Sciences. 2009;1177:66–73. Epub 2009/10/23. doi: 10.1111/j.1749-6632.2009.05039.x. [DOI] [PubMed] [Google Scholar]
- 4.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–98. Epub 2012/09/25. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Payen VL, Porporato PE, Baselet B, Sonveaux P. Metabolic changes associated with tumor metastasis, part 1: tumor pH, glycolysis and the pentose phosphate pathway. Cell Mol Life Sci. 2016;73:1333–48. Epub 2015/12/03. doi: 10.1007/s00018-015-2098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porporato PE, Payen VL, Baselet B, Sonveaux P. Metabolic changes associated with tumor metastasis, part 2: Mitochondria, lipid and amino acid metabolism. Cell Mol Life Sci. 2016;73:1349–63. Epub 2015/12/10. doi: 10.1007/s00018-015-2100-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beadnell TC, Scheid AD, Vivian CJ, Welch DR. Roles of the mitochondrial genetics in cancer metastasis: not to be ignored any longer. Cancer Metastasis Rev. 2018;37:615–32. Epub 2018/12/14. doi: 10.1007/s10555-018-9772-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishikawa K, Koshikawa N, Takenaga K, Nakada K, Hayashi J. Reversible regulation of metastasis by ROS-generating mtDNA mutations. Mitochondrion. 2008;8:339–44. Epub 2008/08/30. doi: 10.1016/j.mito.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science (New York, NY). 2008;320:661–4. Epub 2008/04/05. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 10.Kaipparettu BA, Ma Y, Park JH, Lee TL, Zhang Y, Yotnda P, Creighton CJ, Chan WY, Wong LJ. Crosstalk from non-cancerous mitochondria can inhibit tumor properties of metastatic cells by suppressing oncogenic pathways. PLoS One. 2013;8:e61747. Epub 2013/05/15. doi: 10.1371/journal.pone.0061747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brinker AE, Vivian CJ, Koestler DC, Tsue TT, Jensen RA, Welch DR. Mitochondrial Haplotype Alters Mammary Cancer Tumorigenicity and Metastasis in an Oncogenic Driver-Dependent Manner. Cancer Res. 2017;77:6941–9. Epub 2017/10/27. doi: 10.1158/0008-5472.CAN-17-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paget S The distribution of secondary growths in cancer of the breast. 1889. Cancer metastasis reviews. 1989;8:98–101. Epub 1989/08/01. [PubMed] [Google Scholar]
- 13.Virchow R Cellular pathology. As based upon physiological and pathological histology. Lecture XVI--Atheromatous affection of arteries. 1858. Nutrition reviews. 1989;47:23–5. Epub 1989/01/01. [DOI] [PubMed] [Google Scholar]
- 14.Hart IR, Fidler IJ. Role of organ selectivity in the determination of metastatic patterns of B16 melanoma. Cancer Res. 1980;40:2281–7. Epub 1980/07/01. [PubMed] [Google Scholar]
- 15.Fetterman JL, Zelickson BR, Johnson LW, Moellering DR, Westbrook DG, Pompilius M, Sammy MJ, Johnson M, Dunham-Snary KJ, Cao X, Bradley WE, Zhang J, Wei CC, Chacko B, Schurr TG, Kesterson RA, Dell'italia LJ, Darley-Usmar VM, Welch DR, Ballinger SW. Mitochondrial genetic background modulates bioenergetics and susceptibility to acute cardiac volume overload. Biochem J. 2013;455:157–67. Epub 2013/08/09. doi: 10.1042/BJ20130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feeley KP, Bray AW, Westbrook DG, Johnson LW, Kesterson RA, Ballinger SW, Welch DR. Mitochondrial Genetics Regulate Breast Cancer Tumorigenicity and Metastatic Potential. Cancer Res. 2015;75:4429–36. Epub 2015/10/17. doi: 10.1158/0008-5472.CAN-15-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kesterson RA, Johnson LW, Lambert LJ, Vivian JL, Welch DR, Ballinger SW. Generation of Mitochondrial-nuclear eXchange Mice via Pronuclear Transfer. Bio-protocol. 2016;6. Epub 2016/11/15. doi: 10.21769/BioProtoc.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bayona-Bafaluy MP, Acin-Perez R, Mullikin JC, Park JS, Moreno-Loshuertos R, Hu P, Perez-Martos A, Fernandez-Silva P, Bai Y, Enriquez JA. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 2003;31:5349–55. Epub 2003/09/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welch DR, Neri A, Nicolson GL. Comparison of 'spontaneous' and 'experimental' metastasis using rat 13762 mammary adenocarcinoma metastatic cell clones. Invasion & metastasis. 1983;3:65–80. Epub 1983/01/01. [PubMed] [Google Scholar]
- 20.Poot M `Analysis of intracellular organelles by flow cytometry or microscopy. Current protocols in cytometry. 2001;Chapter 9:Unit 9 4. Epub 2008/09/05. doi: 10.1002/0471142956.cy0904s14. [DOI] [PubMed] [Google Scholar]
- 21.Guo W, Jiang L, Bhasin S, Khan SM, Swerdlow RH. DNA extraction procedures meaningfully influence qPCR-based mtDNA copy number determination. Mitochondrion. 2009;9:261–5. Epub 2009/03/28. doi: 10.1016/j.mito.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shu J, Dolman GE, Duan J, Qiu G, Ilyas M. Statistical colour models: an automated digital image analysis method for quantification of histological biomarkers. Biomed Eng Online. 2016;15:46. Epub 2016/04/29. doi: 10.1186/s12938-016-0161-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cocheme HM, Quin C, McQuaker SJ, Cabreiro F, Logan A, Prime TA, Abakumova I, Patel JV, Fearnley IM, James AM, Porteous CM, Smith RA, Saeed S, Carre JE, Singer M, Gems D, Hartley RC, Partridge L, Murphy MP. Measurement of H2O2 within living Drosophila during aging using a ratiometric mass spectrometry probe targeted to the mitochondrial matrix. Cell Metab. 2011;13:340–50. Epub 2011/03/02. doi: 10.1016/j.cmet.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cocheme HM, Logan A, Prime TA, Abakumova I, Quin C, McQuaker SJ, Patel JV, Fearnley IM, James AM, Porteous CM, Smith RA, Hartley RC, Partridge L, Murphy MP. Using the mitochondria-targeted ratiometric mass spectrometry probe MitoB to measure H2O2 in living Drosophila. Nat Protoc. 2012;7:946–58. Epub 2012/04/21. doi: 10.1038/nprot.2012.035. [DOI] [PubMed] [Google Scholar]
- 25.Ferraro D, Corso S, Fasano E, Panieri E, Santangelo R, Borrello S, Giordano S, Pani G, Galeotti T. Pro-metastatic signaling by c-Met through RAC-1 and reactive oxygen species (ROS). Oncogene. 2006;25:3689–98. Epub 2006/02/08. doi: 10.1038/sj.onc.1209409. [DOI] [PubMed] [Google Scholar]
- 26.Laurila JP, Laatikainen LE, Castellone MD, Laukkanen MO. SOD3 reduces inflammatory cell migration by regulating adhesion molecule and cytokine expression. PloS one. 2009;4:e5786. Epub 2009/06/06. doi: 10.1371/journal.pone.0005786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porporato PE, Payen VL, Perez-Escuredo J, De Saedeleer CJ, Danhier P, Copetti T, Dhup S, Tardy M, Vazeille T, Bouzin C, Feron O, Michiels C, Gallez B, Sonveaux P. A mitochondrial switch promotes tumor metastasis. Cell Rep. 2014;8:754–66. Epub 2014/07/30. doi: 10.1016/j.celrep.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 28.Kaur A, Webster MR, Marchbank K, Behera R, Ndoye A, Kugel CH 3rd, Dang VM, Appleton J, O'Connell MP, Cheng P, Valiga AA, Morissette R, McDonnell NB, Ferrucci L, Kossenkov AV, Meeth K, Tang HY, Yin X, Wood WH 3rd, Lehrmann E, Becker KG, Flaherty KT, Frederick DT, Wargo JA, Cooper ZA, Tetzlaff MT, Hudgens C, Aird KM, Zhang R, Xu X, Liu Q, Bartlett E, Karakousis G, Eroglu Z, Lo RS, Chan M, Menzies AM, Long GV, Johnson DB, Sosman J, Schilling B, Schadendorf D, Speicher DW, Bosenberg M, Ribas A, Weeraratna AT. sFRP2 in the aged microenvironment drives melanoma metastasis and therapy resistance. Nature. 2016;532:250–4. Epub 2016/04/05. doi: 10.1038/nature17392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dikalova AE, Bikineyeva AT, Budzyn K, Nazarewicz RR, McCann L, Lewis W, Harrison DG, Dikalov SI. Therapeutic targeting of mitochondrial superoxide in hypertension. Circulation research. 2010;107:106–16. Epub 2010/05/08. doi: 10.1161/circresaha.109.214601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nazarewicz RR, Dikalova A, Bikineyeva A, Ivanov S, Kirilyuk IA, Grigor'ev IA, Dikalov SI. Does scavenging of mitochondrial superoxide attenuate cancer prosurvival signaling pathways? Antioxidants & redox signaling. 2013;19:344–9. Epub 2013/02/05. doi: 10.1089/ars.2013.5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turrens JF. Mitochondrial formation of reactive oxygen species. The Journal of physiology. 2003;552:335–44. Epub 2003/10/17. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vivian CJ, Brinker AE, Graw S, Koestler DC, Legendre C, Gooden GC, Salhia B, Welch DR. Mitochondrial Genomic Backgrounds Affect Nuclear DNA Methylation and Gene Expression. Cancer Res. 2017;77:6202–14. Epub 2017/07/01. doi: 10.1158/0008-5472.CAN-17-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di A, Mehta D, Malik AB. ROS-activated calcium signaling mechanisms regulating endothelial barrier function. Cell Calcium. 2016;60:163–71. Epub 2016/02/26. doi: 10.1016/j.ceca.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunter KW. Mouse models of cancer: does the strain matter? Nature Rev Cancer. 2012;12:144–9. doi: nrc3206 [pii]; 10.1038/nrc3206 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishikawa K, Hayashi J. A novel function of mtDNA: its involvement in metastasis. Ann N Y Acad Sci. 2010;1201:40–3. Epub 2010/07/24. doi: 10.1111/j.1749-6632.2010.05616.x. [DOI] [PubMed] [Google Scholar]
- 36.Chen Y, Zhang H, Zhou HJ, Ji W, Min W. Mitochondrial Redox Signaling and Tumor Progression. Cancers (Basel). 2016;8. Epub 2016/03/31. doi: 10.3390/cancers8040040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kenny TC, Germain D. mtDNA, Metastasis, and the Mitochondrial Unfolded Protein Response (UPR(mt)). Front Cell Dev Biol. 2017;5:37. Epub 2017/05/05. doi: 10.3389/fcell.2017.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altieri DC. Mitochondrial dynamics and metastasis. Cell Mol Life Sci. 2018, 10.1007/s00018-018-2961-2. Epub 2018/11/12. doi: 10.1007/s00018-018-2961-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreno-Loshuertos R, Acin-Perez R, Fernandez-Silva P, Movilla N, Perez-Martos A, Rodriguez de Cordoba S, Gallardo ME, Enriquez JA. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat Genet. 2006;38:1261–8. Epub 2006/10/03. doi: 10.1038/ng1897. [DOI] [PubMed] [Google Scholar]
- 40.Leary SC, Battersby BJ, Moyes CD. Inter-tissue differences in mitochondrial enzyme activity, RNA and DNA in rainbow trout (Oncorhynchus mykiss). The Journal of experimental biology. 1998;201 (Pt 24):3377–84. Epub 1998/11/18. [DOI] [PubMed] [Google Scholar]
- 41.Rossignol R, Malgat M, Mazat JP, Letellier T. Threshold effect and tissue specificity. Implication for mitochondrial cytopathies. The Journal of biological chemistry. 1999;274:33426–32. Epub 1999/11/24. [DOI] [PubMed] [Google Scholar]
- 42.Kunz WS. Different metabolic properties of mitochondrial oxidative phosphorylation in different cell types--important implications for mitochondrial cytopathies. Experimental physiology. 2003;88:149–54. Epub 2003/01/15. [DOI] [PubMed] [Google Scholar]
- 43.Fan W, Lin CS, Potluri P, Procaccio V, Wallace DC. mtDNA lineage analysis of mouse L-cell lines reveals the accumulation of multiple mtDNA mutants and intermolecular recombination. Genes & development. 2012;26:384–94. Epub 2012/02/22. doi: 10.1101/gad.175802.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jandova J, Shi M, Norman KG, Stricklin GP, Sligh JE. Somatic alterations in mitochondrial DNA produce changes in cell growth and metabolism supporting a tumorigenic phenotype. Biochimica et biophysica acta. 2012;1822:293–300. Epub 2011/11/29. doi: 10.1016/j.bbadis.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parikh VS, Morgan MM, Scott R, Clements LS, Butow RA. The mitochondrial genotype can influence nuclear gene expression in yeast. Science (New York, NY). 1987;235:576–80. Epub 1987/01/30. [DOI] [PubMed] [Google Scholar]
- 46.Burdon RH. Superoxide and hydrogen peroxide in relation to mammalian cell proliferation. Free Radic Biol Med. 1995;18:775–94. Epub 1995/04/01. [DOI] [PubMed] [Google Scholar]
- 47.Sundaresan M, Yu ZX, Ferrans VJ, Irani K, Finkel T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science. 1995;270:296–9. Epub 1995/10/13. [DOI] [PubMed] [Google Scholar]
- 48.Irani K, Xia Y, Zweier JL, Sollott SJ, Der CJ, Fearon ER, Sundaresan M, Finkel T, Goldschmidt-Clermont PJ. Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts. Science. 1997;275:1649–52. Epub 1997/03/14. [DOI] [PubMed] [Google Scholar]
- 49.Sauer H, Wartenberg M, Hescheler J. Reactive oxygen species as intracellular messengers during cell growth and differentiation. Cell Physiol Biochem. 2001;11:173–86. Epub 2001/08/18. doi: 47804. [DOI] [PubMed] [Google Scholar]
- 50.Liu SL, Lin X, Shi DY, Cheng J, Wu CQ, Zhang YD. Reactive oxygen species stimulated human hepatoma cell proliferation via cross-talk between PI3-K/PKB and JNK signaling pathways. Arch Biochem Biophys. 2002;406:173–82. Epub 2002/10/04. [DOI] [PubMed] [Google Scholar]
- 51.Cook-Mills JM, Marchese ME, Abdala-Valencia H. Vascular cell adhesion molecule-1 expression and signaling during disease: regulation by reactive oxygen species and antioxidants. Antioxidants & redox signaling. 2011;15:1607–38. Epub 2010/11/06. doi: 10.1089/ars.2010.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Formentini L, Sanchez-Arago M, Sanchez-Cenizo L, Cuezva JM. The mitochondrial ATPase inhibitory factor 1 triggers a ROS-mediated retrograde prosurvival and proliferative response. Molecular cell. 2012;45:731–42. Epub 2012/02/22. doi: 10.1016/j.molcel.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 53.He J, Xu Q, Jing Y, Agani F, Qian X, Carpenter R, Li Q, Wang XR, Peiper SS, Lu Z, Liu LZ, Jiang BH. Reactive oxygen species regulate ERBB2 and ERBB3 expression via miR-199a/125b and DNA methylation. EMBO reports. 2012;13:1116–22. Epub 2012/11/14. doi: 10.1038/embor.2012.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48:158–67. Epub 2012/10/30. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chae S, Ahn BY, Byun K, Cho YM, Yu MH, Lee B, Hwang D, Park KS. A systems approach for decoding mitochondrial retrograde signaling pathways. Science signaling. 2013;6:rs4. Epub 2013/02/28. doi: 10.1126/scisignal.2003266. [DOI] [PubMed] [Google Scholar]
- 56.Jajoo S, Mukherjea D, Kaur T, Sheehan KE, Sheth S, Borse V, Rybak LP, Ramkumar V. Essential role of NADPH oxidase-dependent reactive oxygen species generation in regulating microRNA-21 expression and function in prostate cancer. Antioxidants & redox signaling. 2013;19:1863–76. Epub 2013/05/21. doi: 10.1089/ars.2012.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raddant AC, Russo AF. Reactive oxygen species induce procalcitonin expression in trigeminal ganglia glia. Headache. 2014;54:472–84. Epub 2014/02/12. doi: 10.1111/head.12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chandel NS. Evolution of Mitochondria as Signaling Organelles. Cell Metab. 2015;22:204–6. Epub 2015/06/16. doi: 10.1016/j.cmet.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 59.Reczek CR, Chandel NS. ROS Promotes Cancer Cell Survival through Calcium Signaling. Cancer Cell. 2018;33:949–51. Epub 2018/06/13. doi: 10.1016/j.ccell.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 60.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–49. Epub 2003/07/05. [DOI] [PubMed] [Google Scholar]
- 61.Hurst DR, Welch DR. Metastasis suppressor genes at the interface between the environment and tumor cell growth. Int Rev Cell Mol Biol. 2011;286:107–80. Epub 2011/01/05. doi: 10.1016/B978-0-12-385859-7.00003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beadnell TC, Scheid AD, Vivian CJ, Welch DR. Roles of the mitochondrial genetics in cancer metastasis: Not to be ignored any longer. Cancer Metastasis Rev. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paget S The distribution of secondary growths in cancer of the breast. Lancet. 1889;1:571–3. [PubMed] [Google Scholar]
- 64.Wallace DC. Mitochondrial genetic medicine. Nat Genet. 2018;50:1642–9. Epub 2018/10/31. doi: 10.1038/s41588-018-0264-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.