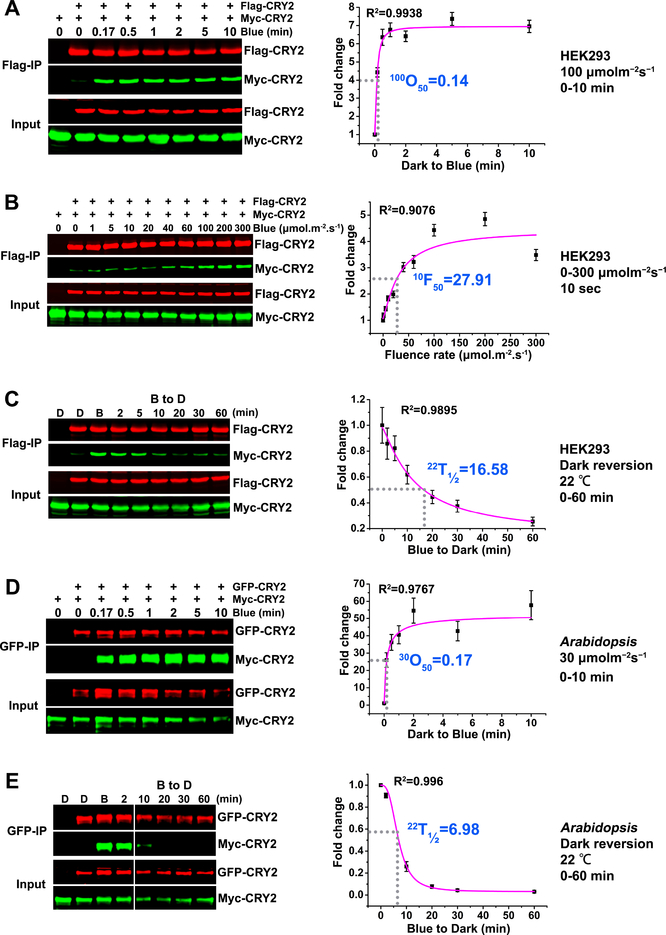
Figure 1. CRY2 photooligomerization ex vivo or in vivo.
(A-C): CRY2 photooligomerization in HEK293 cells (ex vivo).
(A) The kinetics analyses of CRY2 photooligomerization in response to blue light. HEK293 cells were transfected with equal amount of two plasmids encoding Flag-CRY2 or Myc-CRY2. 40 hours after transfection, cells were exposed to blue light of 100 μmolm−2s−1. After 10 seconds to 10 minutes of light treatment, the light-treated and control (left in the dark) cells were killed by lysis buffer. Flag-CRY2 was immunoprecipitated by Flag-agrose gels. The immunoblots were probed by the anti-Flag and anti-Myc antibody, and quantified by the Image Studio Lite software in the Odyssey® CLx System (LI-COR). The Myc-CRY2 (co-IP product) were normalized by the respective Flag-CRY2 (IP product), and presented as Fold Change (FC) by the formula FC = [Myc-CRY2/Flag-CRY2]Blue / [Myc-CRY2/Flag-CRY2]Dark. 100O50 indicates the time (minute) required to reach 50% saturation of CRY2 photooligomerization at the fluence rate of 100 μmolm−2s−1 of blue light.
(B) The fluence-rate response analyses of CRY2 photooligomerization. HEK293 cells transfected with Flag-CRY2 or Myc-CRY2 were exposed to blue light of the indicated fluence rates for 10 seconds. The same analyzation was performed as described in (A). 10F50 indicates the fluence rate of blue light required to achieve 50% CRY photooligomerization after light exposure for 10 seconds.
(C) The dark reversion analyses of CRY2 photooligomerization. HEK293 cells transfected with Flag-CRY2 or Myc-CRY2 were irradiated with 30 μmolm−2s−1 blue light for 5 min before transferring to darkness (at 22°C) for the indicated times. The same analyzation was performed as described in (A-B). 22T1/2 indicates the time (minute) required to reverse 50% of CRY2 photooligomers into monomers at the temperature of 22°C.
(D-E): CRY2 photooligomerization in plants (in vivo).
(D) The kinetics analyses of CRY2 photooligomerization in response to blue light in plants. Seven-day-old Arabidopsis seedlings co-expressing GFP-CRY2 and Myc-CRY2 were grown on petri dishes in the dark, exposed to 30 μmolm−2s−1 blue light, and tissues were collected at the indicated time (minute). GFP-Trap agarose beads were used for immunoprecipitation. The immunoblots were probed by the anti-GFP antibody to detect IP product and anti-Myc antibody to detect Co-IP product, and quantified by the Image Studio Lite software in the Odyssey® CLx System (LI-COR). The Myc-CRY2 (co-IP product) were normalized by the respective GFP-CRY2 (IP product), and presented as Fold Change (FC) by the formula FC = [Myc-CRY2/GFP-CRY2]Blue / [Myc-CRY2/GFP-CRY2]Dark. 30O50 indicates the time (minute) required to reach 50% saturation of CRY2 photooligomerization in plants at the fluence rate of 30 μmolm−2s−1 of blue light.
(E) The dark reversion analyses of CRY2 photooligomerization. Seven-day-old Arabidopsis seedlings co-expressing GFP-CRY2 and Myc-CRY2 were irradiated with 30 μmolm−2s−1 blue light for 5 min before transferring to darkness for the indicated times. 22T1/2 indicates the time (minute) required to reverse 50% of CRY2 photooligomers into monomers.
The data for curve fitting were derived from three technical repeats by performing three independent western blots. The data were analyzed by Logistic Nonlinear Curve Fit method. R2 stands for coefficient of regression. Standard errors are shown.