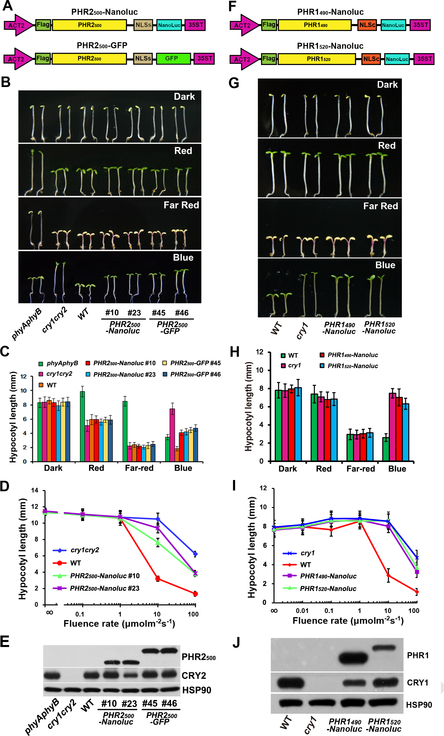
Figure 5. Overexpression of CRY-PHR domain in plants results in reduced sensitivity to blue light.
(A,F) The structures of DNA constructs used for generating CRY2-PHR (A) and CRY1-PHR (F) transgenic lines. PHR2500, CRY2 1–500 aa; PHR1490, CRY1 1–490 aa; PHR1520, CRY1 1–520 aa; NLSs, SV40 nuclear localization signal; NLSc, nuclear localization signal from CRY2 (541–557 aa).
(B,G) Hypocotyl phenotypes of PHR2 (B) and PHR1 (G) transgenic lines. Seedlings with PHR2 or PHR1 overexpressed in wild-type background were grown in the dark, continuous blue light (30 μmol m−2s−1), far-red light (2.5 μmol m−2s−1) and red light (30 μmol m−2s−1) for six days. WT, cry1,cry1cry2 and phyAphyB mutants are used as controls.
(C,H) Hypocotyl measurements of the indicated genotypes shown in (B) and (G). Hypocotyl lengths with standard deviations are shown (n≥20).
(D,I) Fluence rate responses of the hypocotyl inhibition response of PHR2 and PHR1 transgenic lines. Six-day-old seedlings of the indicated genotypes were grown under continuous blue light with fluence rates of 0, 0.1, 1, 10 and 100 μmol m−2s−1. Hypocotyl lengths with standard deviations (n≥20) are shown.
(E,J) Immunoblots showing PHR2 or PHR1 recombinant protein expression levels in transgenic lines. PHR2500, PHR1490 and PHR1520 fusion proteins were detected with anti-Flag antibody. CRY2 and CRY1 proteins were detected by anti-CRY2 or anti-CRY1 antibody, respectively. HSP90 is used as loading control.