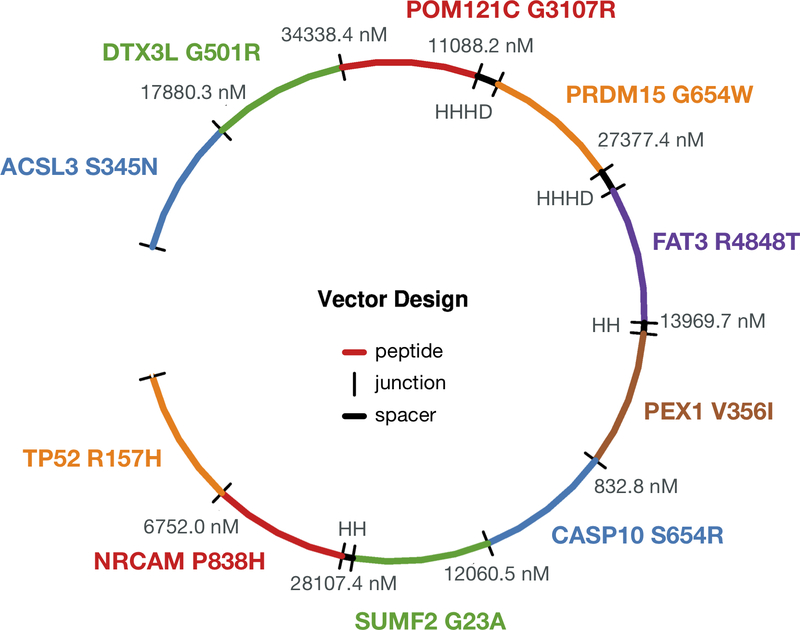
Figure 3: An example pVACvector output showing the optimum arrangement of candidate neoantigens for a DNA-vector based vaccine design.
The figure depicts a circularized DNA insert carrying 10 encoded neoantigenic peptide sequences to be synthesized and encoded/cloned into a DNA plasmid. DNA sequences encoding each peptide are ordered (with use of spacer sequences where needed) to ensure there are no strong-binding junctional epitopes. Each neoantigenic peptide candidate is shown in Blue, Green, Red, Orange, Purple, and Brown. In order to minimize junctional epitope affinity, pVACvector adds spacer sequences where needed. These are depicted in Black, along with the binding affinity value of the junctional epitope. Labels represent the Gene Name and Amino Acid Change for each candidate.