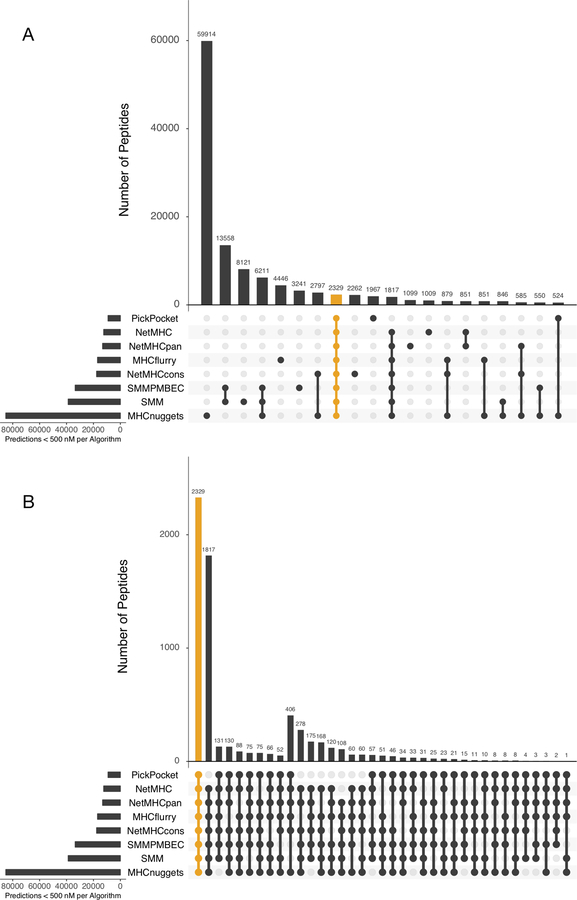
Figure 5: Intersection of peptide sequences predicted by different algorithms (MHCflurry, MHCnuggets, NetMHC, NetMHCcons, NetMHCpan, PickPocket, SMM, and SMMPMBEC):
The y-axis displays the number of overlapping unique neoantigenic peptides predicted for each combination of algorithm depicted on the x-axis. Each collection of connected circles shows the set contained in an exclusive intersection (i.e. the identity of each algorithm), while the light gray circles represent the algorithm(s) that do not participate in this exclusive intersection. (A) Upset plot for the top 20 algorithm combinations ranked by the number of peptides predicted to be a good binder (mutant IC50 score < 500 nM). The combination of all eight algorithms (highlighted orange) ranks 8th highest; (B) Upset plot for algorithm combinations where at least six algorithms agree on predicting a peptide to be a good binder (MT IC50 score < 500 nM). The combination of all eight algorithms (highlighted orange) ranks the highest.