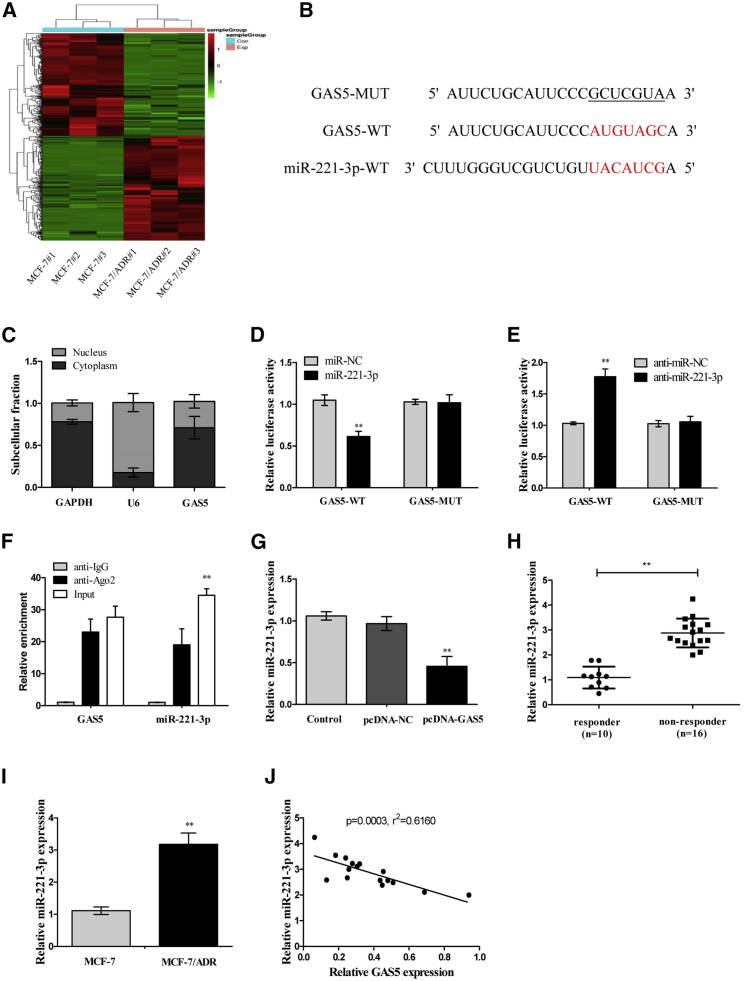
Figure 3.
GAS5 Directly Suppressed miR-221-3p Expression in Breast Cancer Cells
(A) The differently expressed miRNAs between MCF-7 and MCF-7/ADR cells were elucidated by a heatmap (n = 3). (B) Sequence alignment of miR-221-3p with the putative binding sites within the wild-type regions of GAS5. (C) The subcellular location of GAS5 in MCF-7 cells was identified by a subcellular fractionation assay. GAPDH and U6 were used as internal references. (D) Luciferase activity in MCF-7/ADR cells co-transfected with the wipe-type or mutant GAS5 reporters (GAS5-WT or GAS5-MUT) and miR-221-3p mimics or miR-control (Con). (E) Luciferase activity in MCF-7/ADR cells co-transfected with GAS5-WT or GAS5-MUT and anti-miR-221-3p or anti-miR-Con. (F) The GAS5 and miR-221-3p levels isolated from Ago2 and IgG immunoprecipitates derived from MCF-7/ADR cells were examined by quantitative real-time PCR. (G) The miR-221-3p expression in MCF-7/ADR cells transfected with pcDNA-GAS5 was examined by quantitative real-time PCR. (H) The miR-221-3p expression in breast cancer tissues was examined by quantitative real-time PCR. (I) The miR-221-3p expression in MCF-7 and MCF-7/ADR cells was examined by quantitative real-time PCR. (J) Pearson’s analysis shows a negative correlation between GAS5 and miR-221-3p expressions in breast cancer tissues. Data are shown as the mean ± SD (n = 3). *p < 0.05, **p < 0.01 versus respective control.