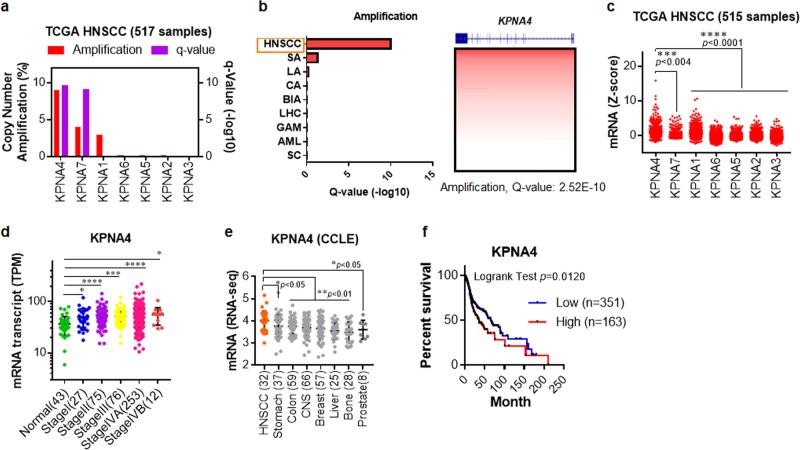
Fig. 1. Profiling of KPNA alteration in HNSCC.
a Analysis of KPNA copy number alteration (CNA) in HNSCC from TCGA (http://cancergenome.nih.gov/). b Summary of KPNA4 amplification across different tumor types from TCGA. SA stomach adenocarcinoma, LA lung adenocarcinoma, CA colon adenocarcinoma, BIA breast invasive adenocarcinoma, LHC liver hepatocellular carcinoma, GM glioblastoma multiform, AML acute myeloid leukemia, SC sarcoma. c The expression of KPNA family transcripts in HNSCC from TCGA. d The expression of KPNA4 in nontumor tissue, and HNSCC samples from Cancer RNA-Seq Nexus (http://syslab4.nchu.edu.tw/). SI, SII, SIII, SIV denote stages I, II, III and IV. e KPNA4 mRNA expression across different types of cancer cells from CCLE (http://portals.broadinstitute.org/ccle/). f High KPNA4 expression (mRNA expression z-Scores (RNA-Seq V2 RSEM) >mean +2.0SD) was associated with poor disease-free survival of HNSCC patients in the TCGA cohorts.