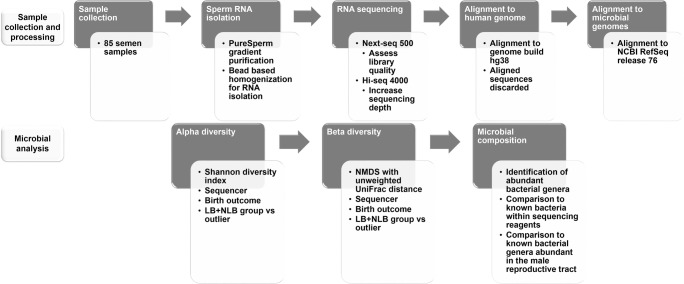
Fig. 1.
Study design. Sample collection and processing is outlined. Sperm RNA from 85 semen samples was isolated and sequenced using a Next-seq 500 and Hi-Seq 4000 sequencer. Sequences were aligned to the human genome then aligned sequences discarded. The unmapped sequences were retained then aligned to the bacterial, viral, and archaeal genomes. Estimated richness of each sample (alpha diversity) employed the Shannon diversity index. Samples were then evaluated as a function of beta diversity (between group richness). With the exception of a single outlier, 84 samples showed the same statistical distribution. Microbial composition was then determined