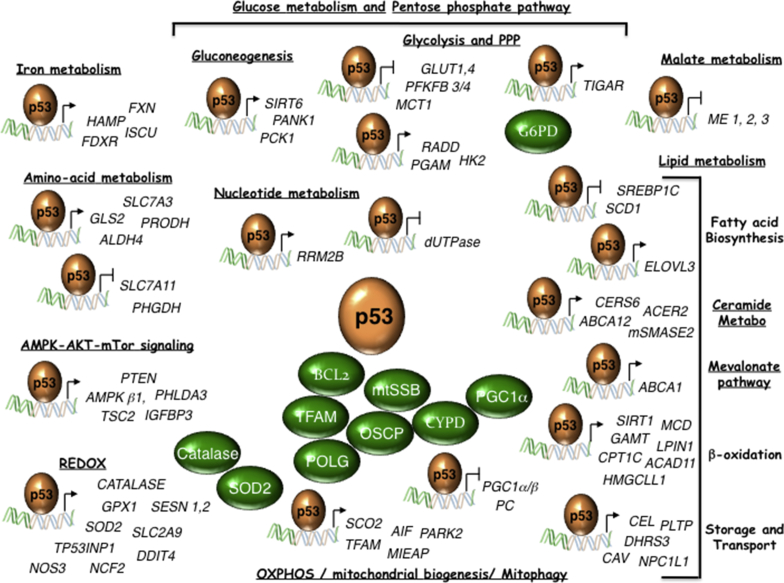
Figure 1.
WT-p53 controls multiple metabolic pathways. p53 direct target genes, and p53-interacting proteins (in green) implicated in metabolism are indicated. ABCA1 (ATP binding cassette subfamily A member 1); ABCA12 (ATP binding cassette subfamily A member 12); ACAD11 (AcylCoA Dehydrogenase Family member 11); ACER2 (Alkaline ceramidase 2); AIF (Apoptosis Inducing Factor); ALDH4 (Aldehyde dehydrogenase 4); AMPKb1 (AMP-activated kinase b1 subunit); BCL2 (B-cell lymphoma 2); Catalase; CAV (Caveolin); CEL (Carboxy Ester Lipase); CERS6 (Ceramide synthetase 6); CPT1C (Carnitine Palmitoyl transferase 1C); CYPD (Cyclophilin D); DDIT4 (DNA Damage inducible transcript 4); DHRS3 (Dehydrogenase reductase 3); dUTPase (deoxyuridine triphosphate nucleotidohydrolase), ELOVL3 (Elongation of very long chain fatty acids-like 3); FDXR (Ferredoxin Reductase); FXN (Frataxin); GAMT (Guanidinoacetate methyltransferase); GLS2 (Glutaminase 2); GLUT1 (Glucose transporter 1); GLUT4 (Glucose transporter 4); GPX1 (Glutathione Peroxidase 1); HAMP (Hepcidin); HK2 (Hexokinase II); HMGCLL1 (3-hydroxymethyl-3-methylglutaryl-CoA lyase like 1); mtSSB (Single Stranded DNA Binding protein 1); IGFBP3 (IGF-Binding protein 3); ISCU (Iron-sulfur cluster assembly enzyme); LPIN1 (Lipin 1); MCD (Malonyl-CoA Decarboxylase); MCT1 (Mono-Carboxylate transporter 1); ME1 (Malic Enzyme 1); ME2 (Malic Enzyme 2); ME3 (Malic Enzyme 3); MIEAP (Mitochondria-eating protein); mSMASE2 (Neutral sphingomyelinase); NCF2 (Neutrophilic cytosolic factor 2); NPC1L1 (Nieman-Pick C1-like 1); NOS3 (Nitric Oxide Synthase 3); OSCP (Oligomycin sensitivity-conferring protein); PANK1 (Panthotenate Kinase 1); PARK2 (Parkin); PC (Pyruvate carboxylase); PCK1 (Phosphoenolpyruvate carboxykinase 1); PFKFB4 (6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 4); PFKFB3 (6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3); PGC1α (Peroxisome proliferator-activated receptor Gamma, Coactivator 1 alpha); PGC1β (Peroxisome proliferator-activated receptor Gamma, Coactivator 1 beta); PGAM (Muscle specific phosphoglycerate mutase); PHGDH (Phosphoglycerate Dehydrogenase); PHLDA3 (Pleckstrin homology-like domain, Family A, member 3); PLTP (Phospholipid Transfer Protein); POLG (Polymerase γ); PRODH (Proline Dehydrogenase); PTEN (Phosphatase and tensin homolog); RRAD (Ras-related associated with Diabetes); RRM2B (Ribonucleotide reductase regulatory subunit M2B); SCD1 (Stearoyl-CoA desaturase 1); SCO2 (Synthesis of cytochrome c oxidase 2); SESN1 (Sestrin 1); SESN2 (Sestrin 2); SIRT1 (Sirtuin 1); SIRT6 (Sirtuin 6); SLC2A9 (Solute carrier family 2 member 9); SLC7A3 (Solute carrier family 7 member 3); SLC7A11 (Solute carrier family 7 member 11); SOD2 (Superoxide Dismutase 2); SREBP1C (Sterol regulatory element binding transcription factor 1); TFAM (Mitochondrial Transcription Factor A); TIGAR (TP53-induced glycolysis and apoptosis regulator); TP53INP1 (Tumor Protein 53-Induced Nuclear Protein 1); TSC2 (TSC Complex subunit 2).