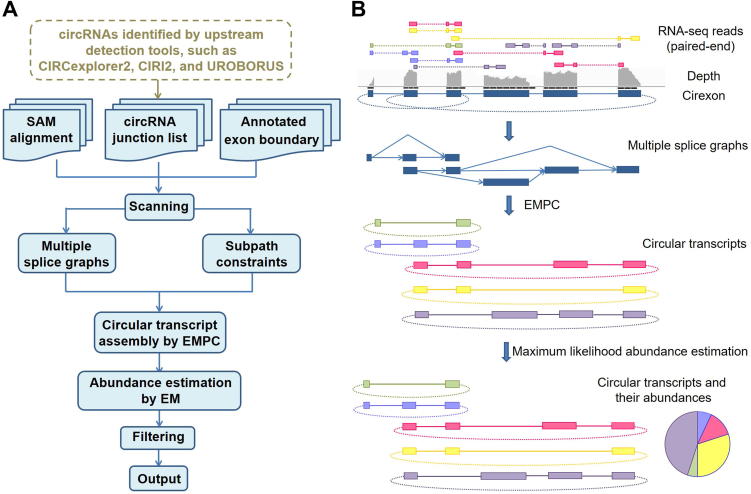
Figure 1.
Schematics of CircAST for circular transcript assembly and quantification
A. The flow diagram of CircAST. B. Visualization of the workflow of CircAST. CircAST begins with a set of paired-end RNA-seq reads that have been mapped to the genome. It then constructs multiple splice graphs with different BSJs in a gene locus and assembles the full-length sequences of circular transcripts with EMPC algorithm. CircAST estimates the abundance of each circular isoform assembled above by using an EM algorithm. Finally, all circular transcripts with full-length sequence assembly and abundance estimation are output in the results. SAM, sequence alignment/map; BSJ, back splice junction; EMPC, extended minimum path cover; EM, expectation maximization.