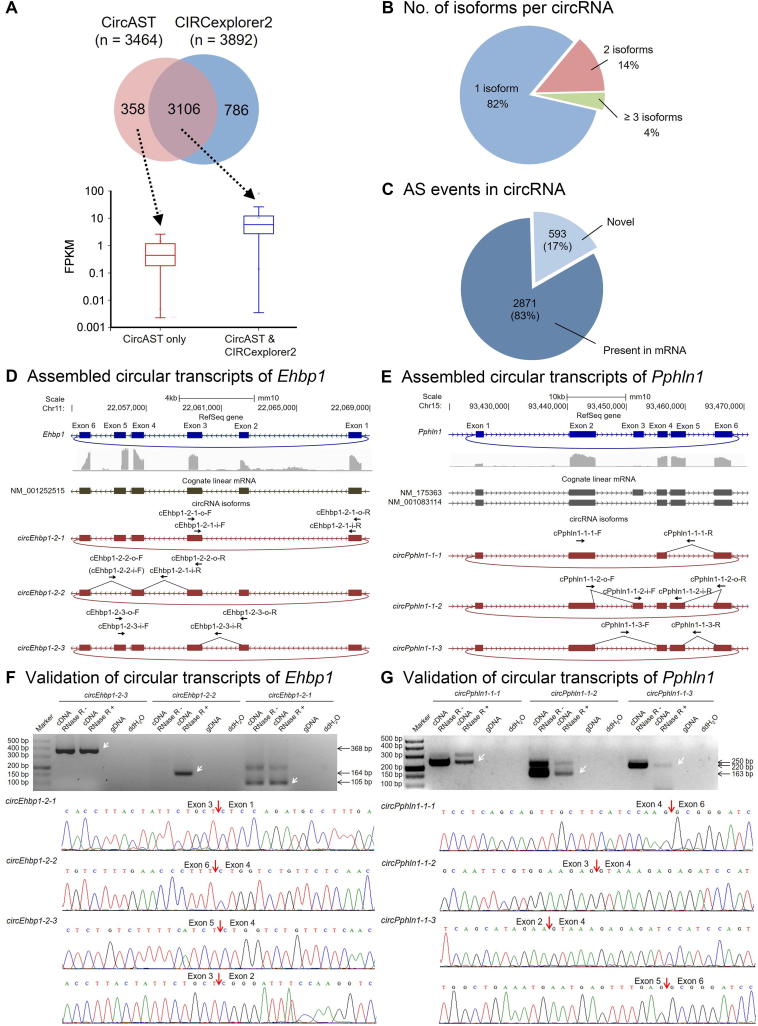
Figure 2.
Circular transcripts assembled by CircAST on adult mouse testis data
A. Venn diagram showing a comparison between the circular isoforms reconstructed by CircAST and CIRCexplorer2 from 2883 mouse testis back-spliced circRNAs (No. of supporting reads ≥10) (Top). Box plots of expression of circRNA isoforms from the Venn diagram, quantified by CircAST in FPKM (Bottom). B. Distribution of isoform number in each back splicing event. C. Comparison of the AS events of circular transcripts with cognate linear transcripts from the same gene locus. D. Visualization of assembled circular transcripts in the gene loci Ehbp1 (Chr11:22,053,432–22,068,506). E. Visualization of assembled circular transcripts in the gene loci Pphln1 (Chr15:93,424,014–93,465,245). For panels D and E, PCR primers are annotated together with the parental gene structure and cognate linear mRNA model. F. RT-PCR and Sanger sequencing for all the circular transcripts in the gene loci Ehbp1 (Chr11:22,053,432–22,068,506). G. RT-PCR and Sanger sequencing for all the circular transcripts in the gene loci Pphln1 (Chr15:93,424,014–93,465,245). For panels F and G, white arrows point to the isoform in the RNase R-treated sample with the correct product size, which was later subjected to Sanger sequencing. There are three isoforms for both circRNAs, in which circEhbp1-2-2 and circPphln1-1-1 were predicted by only CircAST but missed by CIRCexplorer2. The primer sequences are provided in Table S1. AS, alternative splicing.