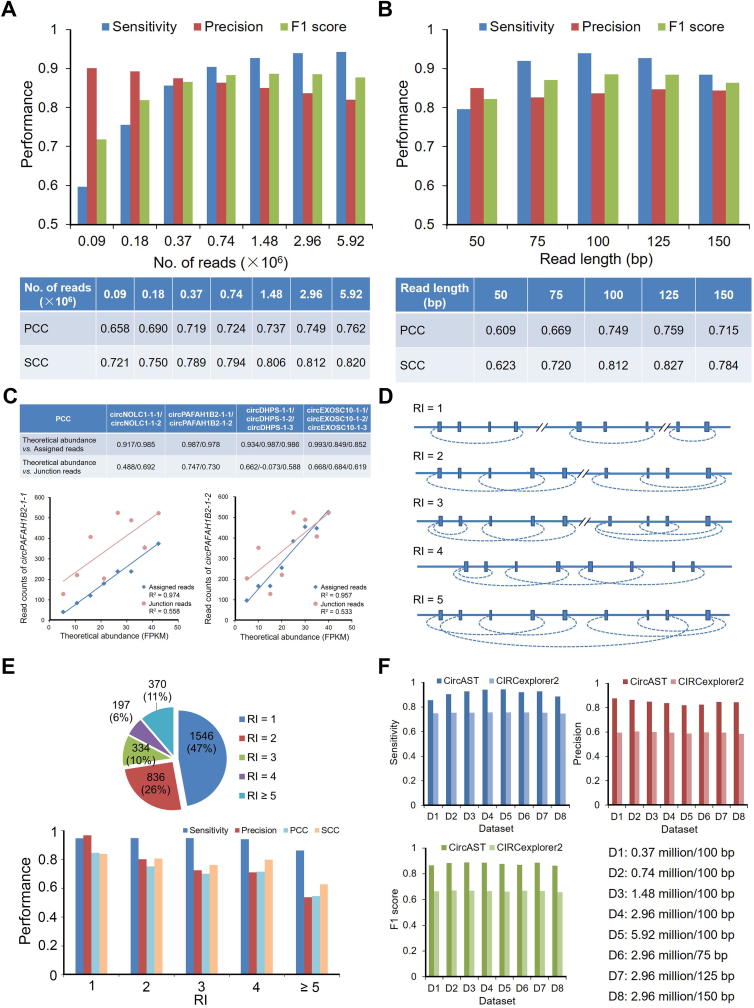
Figure 3.
Performance of CircAST on simulated datasets
A. The sensitivity, precision, and score of full-length assembly by CircAST (upper), and PCC and SCC (lower) between theoretical and estimated absolute expression levels across different circular transcripts assembled by CircAST in simulated datasets with different sequencing depths. B. The sensitivity, precision, and score of full-length assembly by CircAST (upper), and Pearson and Spearman correlation coefficients (lower) between theoretical and estimated absolute expression levels across different circular transcripts assembled by CircAST in simulated datasets with different read lengths. C. PCC between the theoretical and estimated relative expression levels by CircAST for four back-spliced loci with two or more isoforms due to AS in simulated datasets. Correlation plots between the theoretical abundance and calculated read counts from junction reads or assigned reads by CircAST are shown for circPAFAH1B2-1-1 and circPAFAH1B2-1-2. D. Schematics of RI of equivalence classes to indicate the complexity of circRNAs due to the presence of overlapping regions. E. RI distribution of equivalence classes and effects of RI on the accuracy of circular transcript assembly and abundance estimation by CircAST. F. Comparison of the sensitivity, precision, and score between CircAST and CIRCexplorer2 on simulated datasets with different read lengths and sequencing depths. PCC, Pearson correlation coefficient; SCC, Spearman correlation coefficient; RI, relation index.