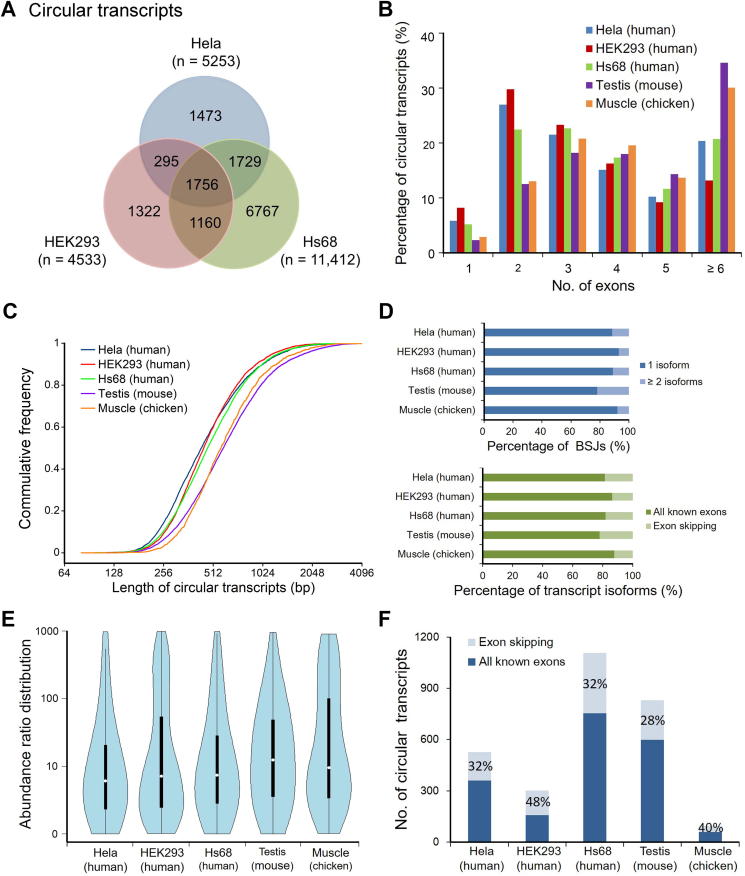
Figure 4.
Analysis of circular transcripts assembled by CircAST in three human cell lines, mouse testis, and chicken muscle
A. Venn diagram depicting the diversity of circular transcripts assembled by CircAST among the three human cell lines HeLa, HEK293, and Hs68. B. The distribution of exon number in circular transcripts in different cell lines and tissues. C. Length distribution of circular transcripts in different cell lines and tissues. D. Distribution of circular isoform number in each back splicing event, and the percentage of transcript isoforms containing all known exons between back splice sites in different cell lines and tissues. E. The abundance ratio distribution of the top two most abundant circular isoforms from circRNAs with multiple isoforms in each back splice site. F. For the most abundant isoform in each circRNA with multiple isoforms, there is a large proportion (28%–48%) of exon skipping events.