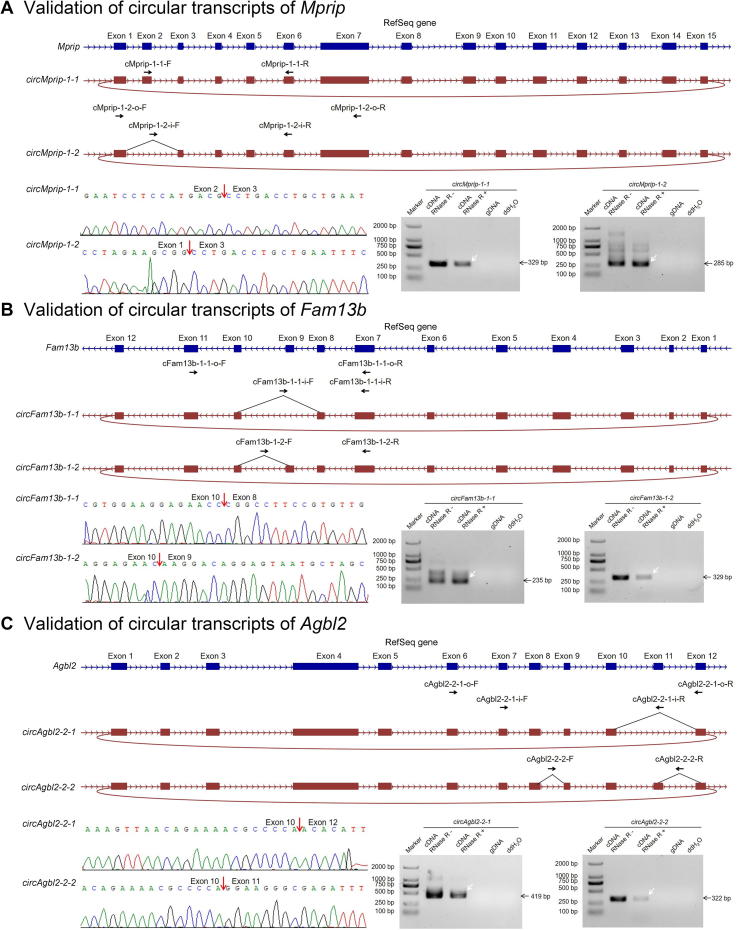
Supplementary Figure S6-1.
Validation of the long circRNA isoforms in different gene loci predicted by CircAST The schematic models of circRNA isoforms of Mprip (Chr11: 59,741,120–59,771,702) (A), Fam13b (Chr18: 34,443,901–34,465,220) (B), Agbl2 (Chr2: 90,791,460–90,813,352) (C), Sbno1 (Chr5: 124,384,467–124,414,527) (D), Bptf (Chr11: 107,043,633–107,077,798) (E), Helz (Chr11: 107,592,706–107,649,340) (F), March6 (Chr15: 31,478,295–31,509,822) (G), Zfp638 (Chr6: 83,934,949–83,972,283) (H), and Ascc3 (Chr10: 50,690,115–50,767,507) (I) are provided separately (top rows). The RT-PCR results for each circRNA isoform and the corresponding Sanger sequencing data showing correct specific splicing sites (bottom rows) are provided for validation. J.Gapdh was used as a negative control.