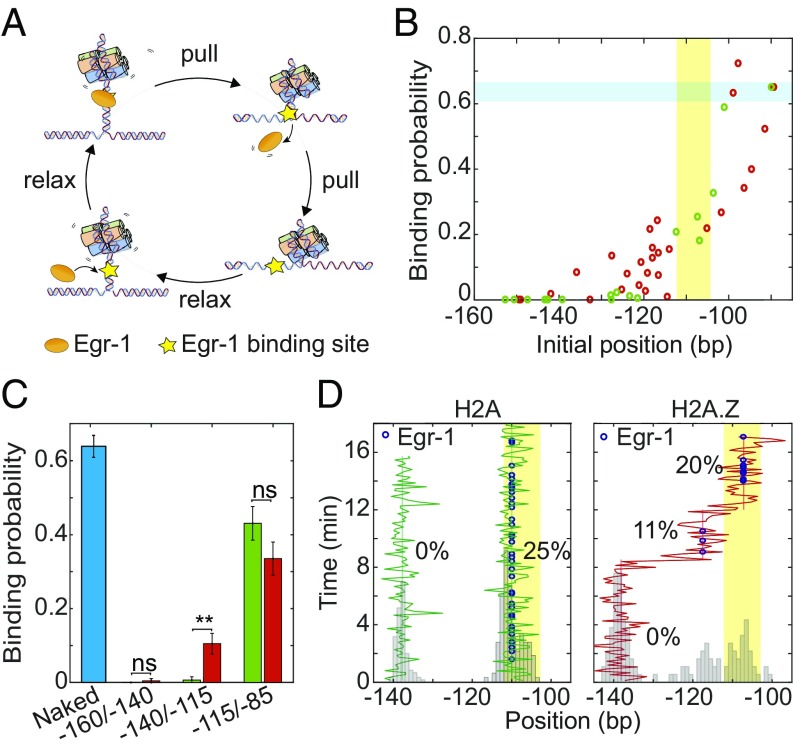
Fig. 3.
Nucleosome diffusion facilitates TF binding. (A) Experimental scheme for the repetitive probing of nucleosome positioning in the presence of Egr-1. (B) Calculated binding probability for Egr-1 as a function of nucleosome initial position, for experiments longer than 7 min. Data are shown as the result of individual experiments; nH2A = 25 and nH2A.Z = 20. The Egr-1 binding site is highlighted in yellow. The binding probability of Egr-1 to naked DNA (highlighted in blue) is shown for comparison. (C) Average binding probability for H2A (green) and H2A.Z (red) nucleosomes whose initial position is located between −160/−140 (nH2A = 3; nH2A.Z = 4), −140/−115 (nH2A = 5; nH2A.Z = 15), and −115/−85 (nH2A = 12; nH2A.Z = 6). Shown as mean ± SEM. The binding probability of Egr-1 to naked DNA (blue) is shown for comparison (n = 262; eight experiments). (D) Repositioning trajectory of two H2A (Left, green) and a single H2A.Z (red, Right) nucleosomes. Histograms of the positions are shown in gray. The location of the Egr-1 binding site is marked in yellow. The blue dots represent detection of a bound Egr-1 in the corresponding cycle, and the numbers, the percent of the probing cycles where a protein was bound, for different mean positions of the nucleosome.