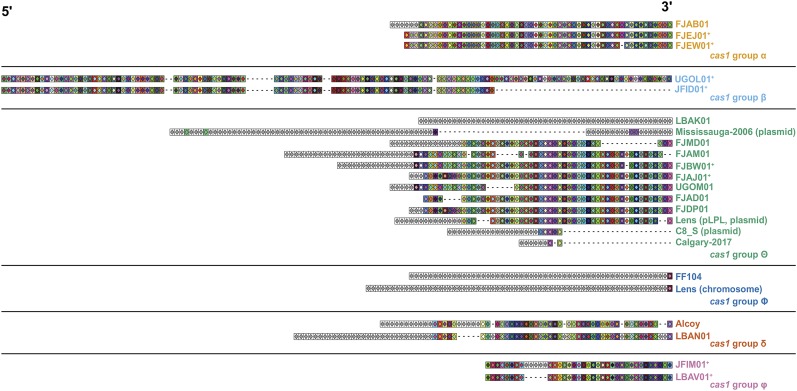
Figure 2.
CRISPR array analysis suggests that spacer acquisition and spacer loss has contributed to array diversification in L. pneumophila type I-F systems. L. pneumophila isolates were subjected to a CRISPRStudio analysis to look at the spacer composition of their CRISPR arrays. Gray boxes denote unique spacers, colored boxes denote shared spacers and a dashed line denotes spacer loss. The isolate color coding scheme is based on the cas1 grouping from Figure 1. Isolates denoted with a “+” within the same cas gene group have 100% nucleotide identity in their core genomes. Three strains have systems that reside on contigs previously annotated as plasmids (str. Lens, str. Mississauga-2006, and str. C8_S). Notably, an additional six strains (FJAD01, FJAJ01, FJAM01, FJBU01, FJBW01 and FJMD01) have systems that reside on contigs with overlapping sequence (78-89 nt) on either end, suggesting that they may also reside on a plasmid or other mobile element.