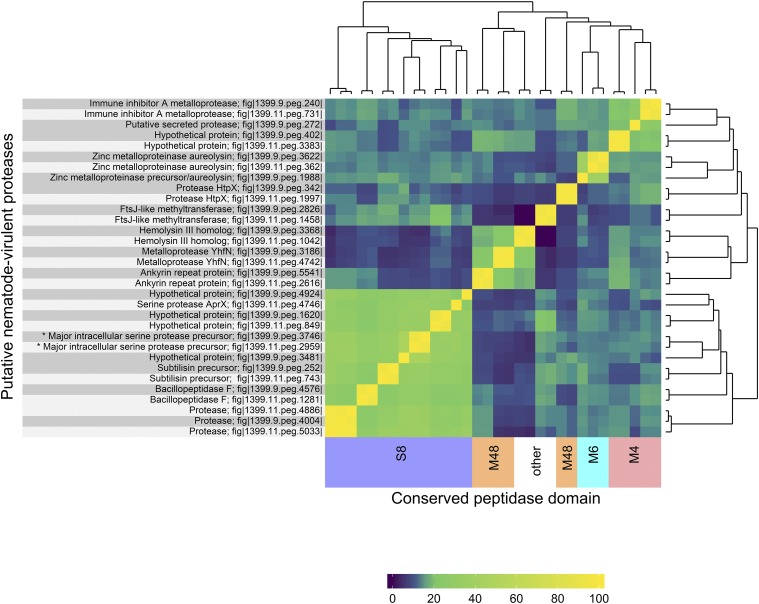
Figure 3.
Clustering of putative nematode-virulent proteases found in Bacillus firmus I-1582 (light-gray) and Bacillus sp. ZZV12-4809 (dark-gray) genomes. All-against-all comparison of amino-acid sequences is based on percent (%) identity values calculated with Clustal Omega and presented in a heatmap, ranging from lower (violet) to higher sequence identity (green-yellow). A dendrogram was created using hierarchical clustering by Euclidean distance. The identities of analyzed proteins are given as genomic annotations together with PATRIC IDs (left-hand margin). Colored highlights (bottom margin) represent clustering of sequences according to the conserved peptidase family domains: S8 (blue), M4 (magenta), M6 (cyan), M48 (orange), or other (no color). Asterisks (*) denote high-similarity protein sequences to the nematicidal protease Sep1 from B. firmus DS1.