Figure 1. Flow chart for the integrated use of the tools.
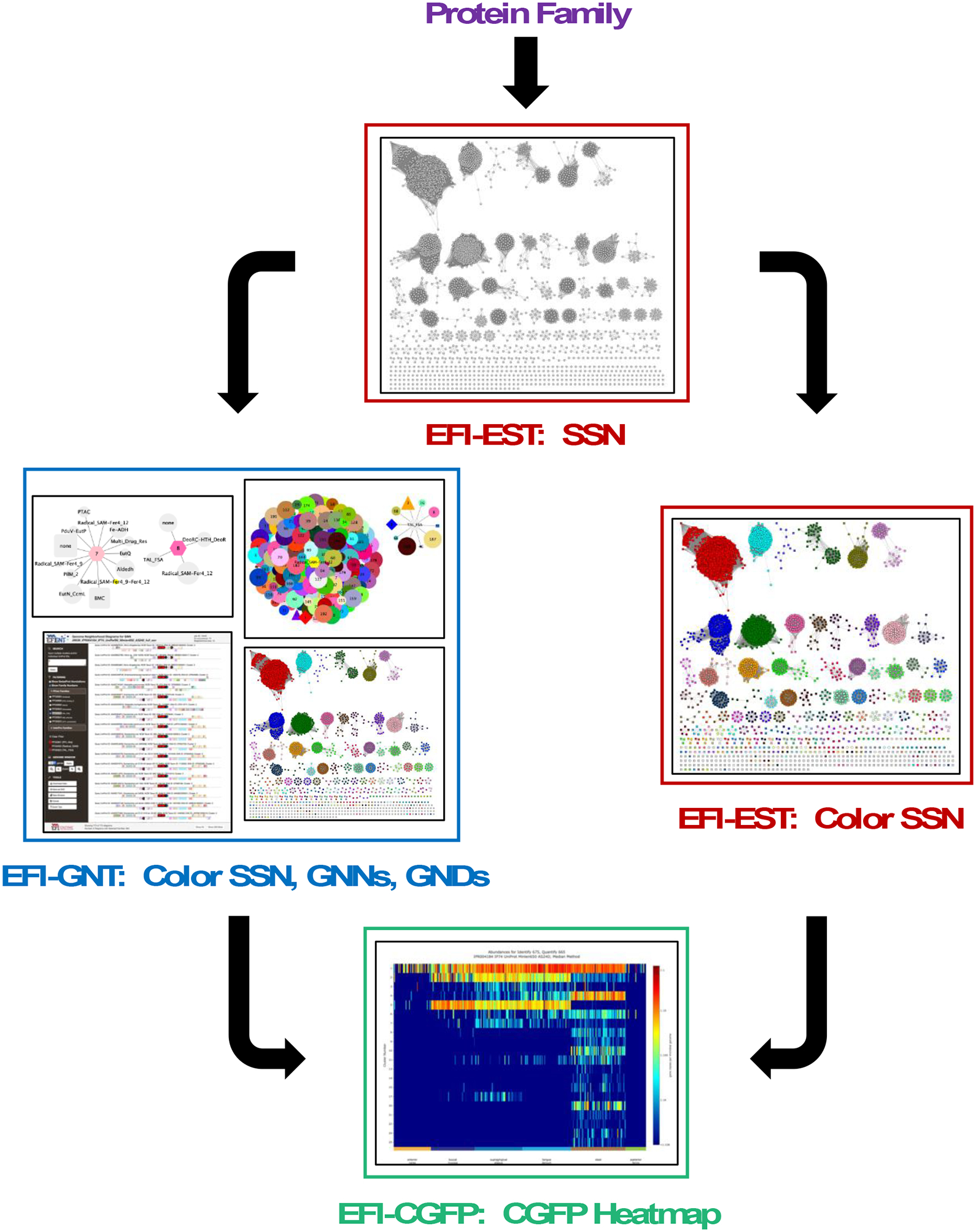
The input for the EFI-EST tool is a user-specified protein family, in this manuscript, the GRE superfamily (IPR004184); the output from EFI-EST is an SSN. The clusters in the SSN can be assigned unique colors and numbers using the Color SSNs utility (right path) to generate a colored SSN. The SSN from EFI-EST also can be used as the input for the EFI-GNT tool (left path) that generates 1) a colored SSN that also contains genome neighborhood information 2) two genome neighborhood networks (GNNs) that summarize genome context, and 3) genome neighborhood diagrams (GNDs) that allow visualization of the genome neighborhoods for each bacterial, archaeal, and fungal protein in each SSN cluster of the input SSN. The colored SSN from either the Color SSNs utility or EFI-GNT is used as input for EFI-CGFP to map metagenome abundance to SSN clusters; the output includes a colored SSN with information about family specific sequence markers and their metagenome abundance and a heatmap that provides a visual summary of metagenome abundance for each SSN cluster and singleton.
