Figure 3. UniRef90 SSNs for IPR004184 generated at several alignment scores illustrating the procedure to obtain isofunctional (orthologous) clusters.
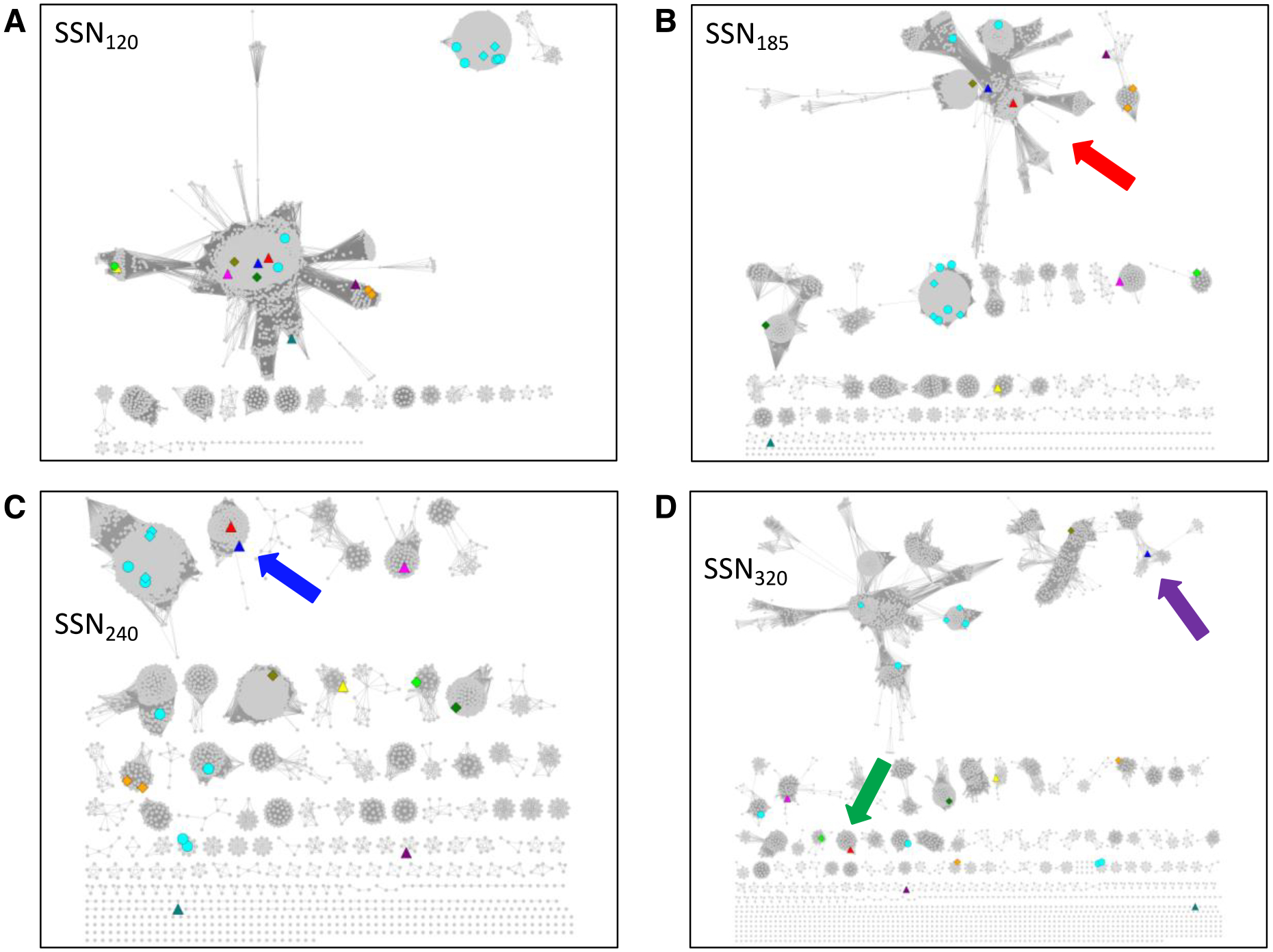
Panel A, alignment score threshold of 120; the eleven characterized functions are not segregated. Panel B, alignment score threshold of 185; the dehydratase functions are located in the same cluster (red arrow), highlighting the ability of SSNs to associate related functions. Panel C, alignment score threshold of 240; ten of the eleven characterized functions are in separate clusters; glycerol dehydratase and 1,2-propanediol dehydratase are in the same cluster (blue arrow; same reactions using similar substrates). Panel D, alignment score threshold of 320; the glycerol dehydratase (magenta arrow) and 1,2-propanediol dehydratase (green arrow) functions are segregated. Nodes are colored as described in the text: PFL, cyan; choline trimethylamine lyase, green; trans-4-hydroxy-L-protline dehydratase, olive; 4-hydroxyphenylacetate decarboxylase, orange; benzylsuccinate synthase; lime; glycerol dehydratase, blue; 1,2-propanediol dehydratase, red; isethionate sulfite lyase, magenta; alkylsuccinate synthase, yellow; indoleacetate decarboxylase, purple; and phenylacetate decarboxylase, teal.
