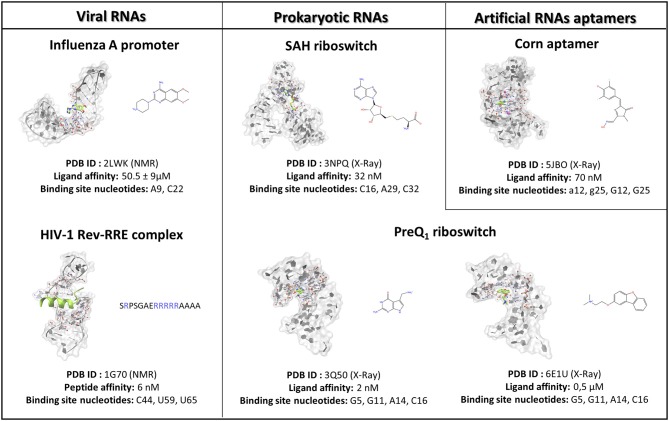
Figure 1.
The case studies selected for the SuMD methodological validation are herein summarized and subdivided into RNA of viral origin, prokaryotic origin, or artificially engineered aptamers. For each complex investigated, the three-dimensional structure is depicted, representing with a green color the reference ligand, together with the nucleobases selected to define the binding site position in the SuMD simulations. Finally, the chemical structures of each ligand are reported, along with the experimental datum of binding affinity. In the case of the peptide, the primary sequence is reported, highlighting the basic residues constituting the arginine reach motif (ARM) in a blue color.