Figure 1.
MeRIP-Seq Analysis Unveils the m6A Profiles in the OSCC Cells
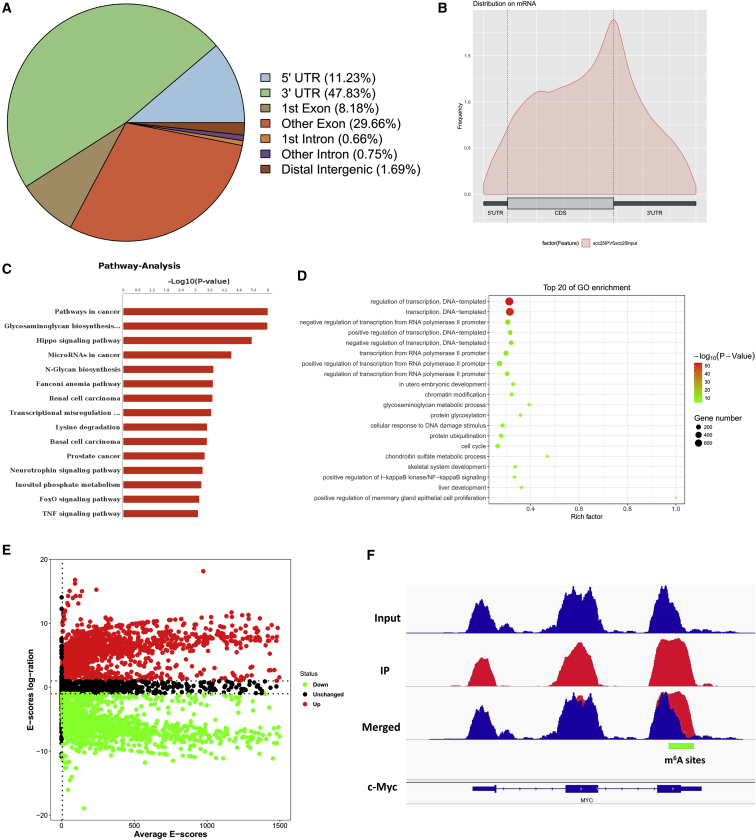
(A) m6A-RNA immunoprecipitation sequencing (MeRIP-seq) was performed. Metagene profile illustrated the m6A distribution in 5′ untranslated region (UTR), coding sequences (CDS), stop codon, and 3′ UTR. (B) Metagene profile of m6A distribution showed the m6A peaks in the stop codon, including the CDSs and 3′ UTRs. (C) Pathway analysis showed the potential pathway for the OSCC. (D) Gene Ontology (GO) analysis showed the target manners for the OSCC. (E) Volcano plot for MeRIP-seq analysis showed the different expression of transcripts, including upregulated and downregulated transcripts. (F) The Integrative Genomics Viewer (IGV) tool revealed the m6A peak distribution c-Myc mRNA.