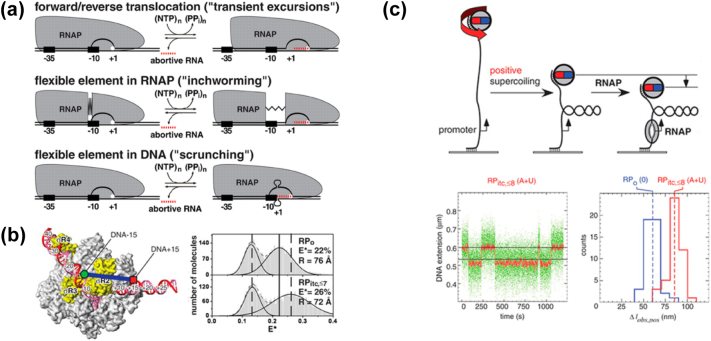
Fig. 3.
A) Schematic of the three proposed models for mechanism of initial transcription: (top), transient excursions; (middle) inchworming; (bottom) scrunching.; B) smFRET study showing scrunching during initial transcription; single molecule assay showing increasing FRET efficiency (decrease in distance) for dyes in positions − 15 (Cy3b) and + 15 (Alexa647) of a lacCONS promoter. Subpanels show E ∗ histograms of open complex (RPo) and initial transcribing complexes with up to 7-nt RNA (RPITC, ≤ 7). The histograms show distributions of free DNA (lower- E ∗ species) and the RNA polymerase (RNAP)–DNA complexes. An increase in FRET efficiency indicates a decrease in distance in going from RPo to RPITC and supports the scrunching model for initial transcription; (from Kapanidis et al, Science 2006 [64], used with permission). C) magnetic tweezer study showing 1 bp scrunching per nucleotide addition cycle during initial transcription. (top) the end-to-end extension of a mechanically stretched, negatively supercoiled or positively supercoiled single DNA molecule containing a single promoter is monitored. Unwinding of n turn of DNA by RNAP result in the compensatory loss of n negative supercoils or gain of n positive supercoils and a readily detectable movement of the bead; (bottom) magnetic tweezers data showing scrunching of DNA during initial transcription (adapted from Revyakin et al, Science, 2006 [65]; used with permission).