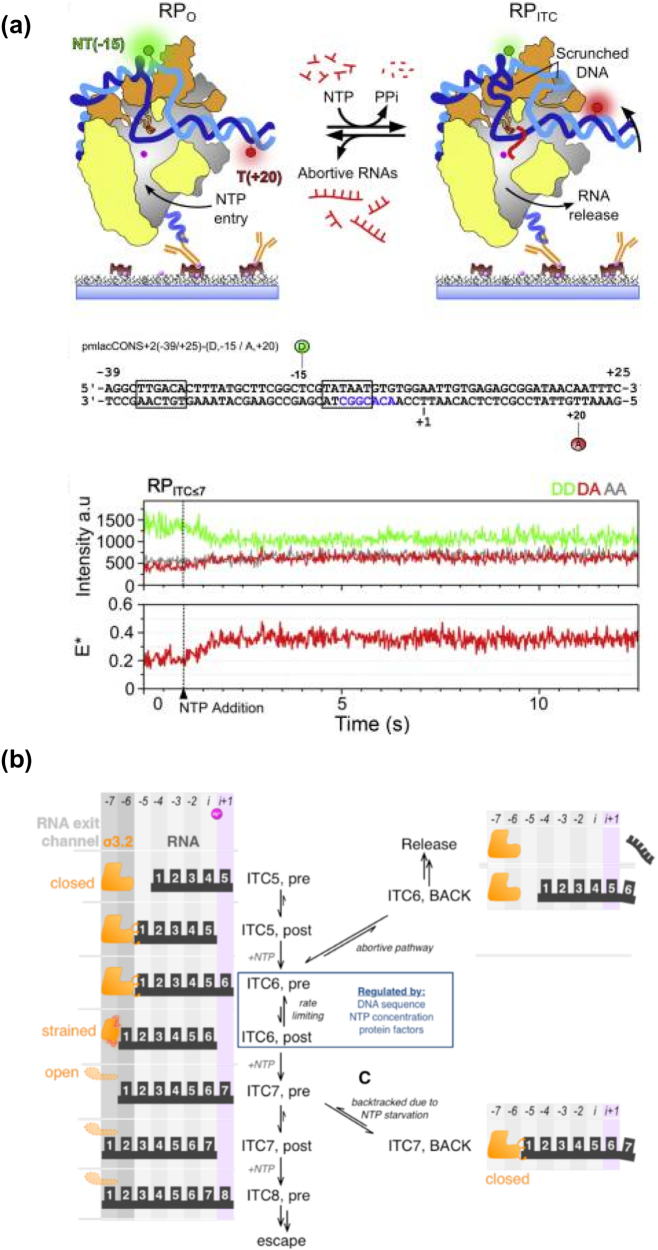
Fig. 4.
A) smFRET assay showing pausing in initial transcription; (top): left, RPo; right, initial transcribing complex (ITC). Donor is in green; acceptor in red; σ70 in orange; RNAP in grey, except for the β subunit (omitted for clarity) and regions protruding from the cut-away plane (in yellow); template strand in blue; non-template strand in teal; nascent RNA in red; and RNAP active site in pink. The penta-His antibody anchors RPo to the surface. The initial FRET efficiency is low; upon NTP addition, scrunching moves the acceptor closer to the donor, increasing FRET efficiency; (middle): lacCONS DNA fragment for FRET assay; the − 10/−4 pre-melted region is in blue; (bottom): time trace showing an increase to E ∗ ∼ 0.37 upon adding 80 μM UTP and GTP to form RPITC≤7. The NTP addition point is marked with a dashed line. Frame time: 20 ms. DD trace (green trace, top), donor emission upon donor excitation; DA trace (red trace, top), acceptor emission upon donor excitation; AA trace (grey trace, top), acceptor emission upon acceptor excitation. DD and DA are used for calculating apparent FRET efficiency E ∗; B) Model for pausing in initial transcription showing the different elements in the RNAP-promoter complex in play; (top): productive path for initial transcription. Coloured columns show translocational registers adopted by growing RNA (in black). Binding site for incoming NTP is in light purple; σ3.2 loop is shown in three putative conformations (in orange). The translocational equilibrium for RPITC6 is controlled by several regulatory factors that modulate the lifetime of paused states arising from a pre-translocated RPITC6; (middle): abortive path for initial transcription, branching from the pre-translocated RPITC6 state of the productive path; (bottom): path for the formation of stable backtracked scrunched states, branching from the pre-translocated RPITC6 state of initial transcription during NTP starvation that limits RNA synthesis to 7 nt in length. (adapted from Duchi et al, Mol Cell, 2016 [69]; used with permission).