Figure 2.
Cohesin-SA2 Facilitates the Establishment of Polycomb-Repressed Regions in mESCs
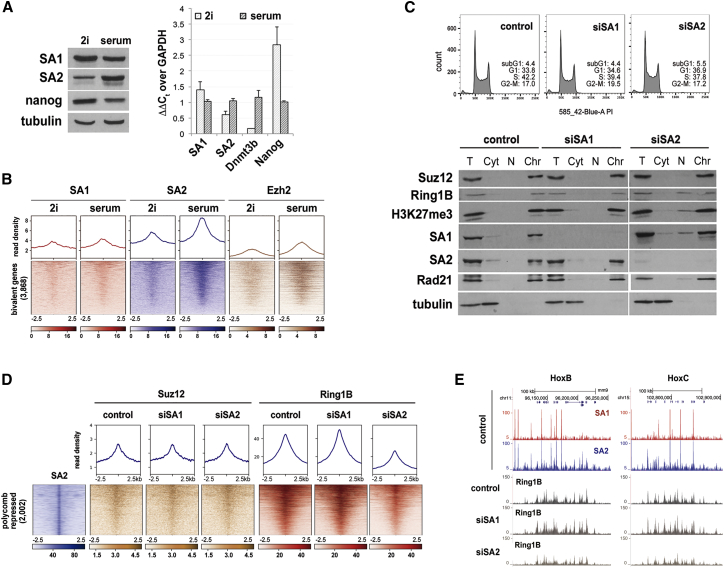
(A) Comparison of protein (left) and mRNA levels (right) of SA1, SA2, and Nanog in mESCs growing in 2i and serum conditions. Tubulin, loading control for immunoblot. For qRT-PCR (right), data represent mean and SD from three independent experiments. Low Dnmt3b expression is a feature of mESCs cultured in 2i (Leitch et al., 2013).
(B) Distribution of SA1, SA2, and PRC2 protein Ezh2 assayed by ChIP-seq around bivalent gene promoters defined by Mas et al. (2018) in mESCs grown in the indicated conditions. Data for Ezh2 are from Marks et al. (2012).
(C) Cell cycle profiles of mock-depleted (control) and SA1- or SA2-depleted mESCs (top). The presence of cohesin and Polycomb proteins on chromatin after downregulation of SA1 or SA2 was assessed by chromatin fractionation. T, total cell extract; Cyt, cytosol; N, nuclear soluble; Chr, chromatin.
(D) Read density plots (top) and read heatmap (bottom) comparing the distribution of Suz12 and Ring1B around the 2,002 SA2-only positions containing Polycomb, described in Figure 1A, in mock-depleted (control), SA1-depleted (siSA1), and SA2-depleted (siSA2) cells.
(E) UCSC Genome Browser image of the HoxB and HoxC loci showing ChIP-seq read distribution for SA1 and SA2 in mESCs as well as for Ring1B in control and SA1- or SA2-depleted cells.