Figure 3.
Cohesin Variants Make Different Contributions to Genome Architecture in mESCs
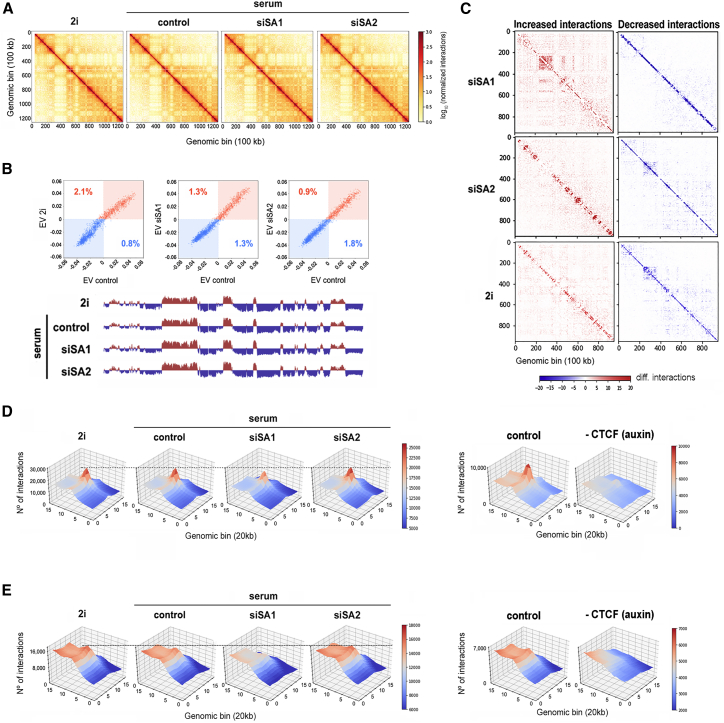
(A) Vanilla-normalized Hi-C matrices for chromosome 17 at 100 kb resolution in mESCs treated as indicated. All analyses in this figure use data merged from two replicates.
(B) Scatterplot of eigenvectors (EVs) of the intrachromosomal interaction matrices indicated in the axis. Numbers within the plot show the percentage of bins that changed compartment. The first eigenvector for chromosome 17 at 100 kb resolution is shown below the plots. Blue and red signals correspond to B and A compartments, respectively.
(C) Matrices showing increased (red) and decreased (blue) interactions in chromosome 17 when comparing siSA1 (top), siSA2 (middle), or 2i-growing cells with serum-growing mock-depleted (control) cells. Similar results were obtained in the analysis of individual replicates and additional chromosomes (Figure S2).
(D) Three-dimensional interaction meta-plots showing contact strength in long (>500 kb) cohesin-mediated loops previously defined by Hi-ChIP (Mumbach et al., 2016) in the different conditions. For comparison, the effect of CTCF depletion was also analyzed using data previously generated in mESCs carrying auxin-inducible degron (AID)-CTCF (Nora et al., 2017).
(E) Meta-analysis of loop strength as in (D) but using borders of high-resolution TADs defined by Bonev et al. (2017). Replicates were also analyzed separately (Figure S3).