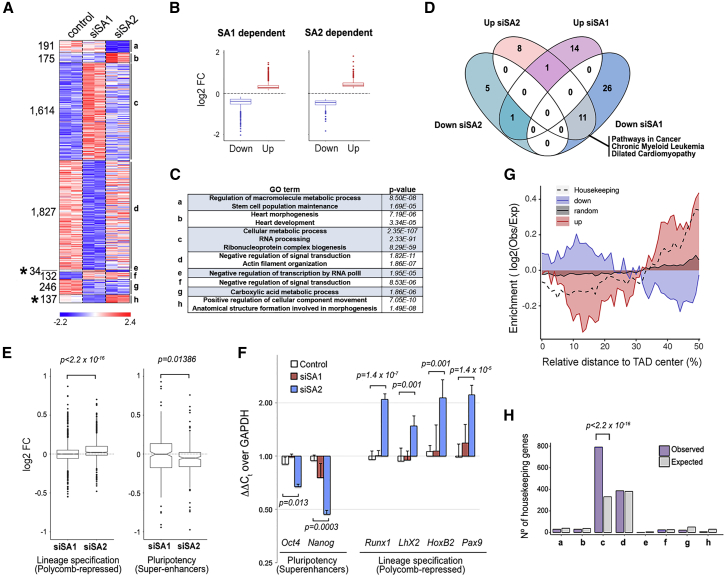
Figure 6.
Distinct Effects of Cohesin Variants on Transcription
(A) Heatmaps showing differentially expressed genes in the indicated conditions (two independent replicates). Eight different clusters of genes were revealed according to their response to SA1 or SA2 depletion (a–h). Numbers on the left indicate the number of genes in each cluster. See also Tables S3 and S4.
(B) Boxplots showing changes in expression for upregulated and downregulated genes in serum-growing mESCs depleted from SA1 or SA2 compared with control. Boxes in (B) and (E) represent interquartile range (IQR); the midline represents the median; whiskers are 1.5 × IQR; and individual points are outliers.
(C) Some of the most significantly enriched GO terms in each cluster defined in (A) are shown. p values were calculated with a Fisher’s exact test and corrected by FDR (< 0.05).
(D) Venn diagram showing the overlap between the KEGG pathways obtained by GSEA of genes deregulated after SA1 or SA2 depletion. See also Table S5.
(E) Boxplots comparing changes in expression after SA1 or SA2 depletion for Polycomb-repressed (left) and super-enhancer-dependent (right) genes. Polycomb-repressed genes (n = 1,008) were defined as those having Ring1B at their promoter and fragments per kilobase of transcript per million mapped reads (FPKM) < 1 in the control condition. Super-enhancer-dependent genes (n = 278) were those defined by Novo et al. (2018). Statistical significance was calculated using a Wilcoxon signed rank test.
(F) Changes in expression of some Polycomb-repressed and super-enhancer-dependent genes were assessed using qRTPCR and normalized to levels of the housekeeping gene GAPDH. Data are mean and SD from at least three independent experiments. A Student’s t test was used to assess statistical significance.
(G) Plot showing the relative distribution of genes deregulated only in the siSA1 condition (up or down) as well as the 4,781 mouse housekeeping genes defined by Li et al. (2017) along TADs in mESCs. Black line represents the distribution of genes not affected by SA1 depletion.
(H) Histogram showing the observed and expected distribution of the housekeeping genes among the eight groups of cohesin regulated genes defined in (A). Statistical significance was assessed using a Fisher’s exact test.