Abstract
The construction of scaffolds and subsequent incorporation of cells and biologics have been widely investigated to regenerate damaged tissues. Scaffolds act as a template to guide tissue formation, and their characteristics have a considerable impact on the regenerative process. Whereas many technologies exist to induce specific two-dimensional (2D) morphologies into biomaterials, the introduction of three-dimensional (3D) micromorphologies into individual pore walls of scaffolds produced from biological molecules such as collagen poses a challenge. We here report the use of dicarboxylic acids to induce specific micromorphologies in collagen scaffolds and evaluate their effect on cellular migration and differentiation. Insoluble type I collagen fibrils were suspended in monocarboxylic and dicarboxylic acids of different concentrations, and unidirectional and random pore scaffolds were constructed by freezing and lyophilization. The application of various acids and concentrations resulted in variations in 3D micromorphologies, including wall structure, wall thickness, and pore size. The use of dicarboxylic acids resulted in acid-specific micromorphologies, whereas monocarboxylic acids did not. Dicarboxylic acids with an odd or even number of C-atoms resulted in frayed/fibrillar or smooth wall structures, respectively, with varying appearances. The formation of micromorphologies was concentration-dependent. In vitro analysis indicated the cytocompatibility of scaffolds, and micromorphology-related cell behavior was indicated by enhanced myosin staining and myosin heavy chain gene expression for C2C12 myoblasts cultured on scaffolds with frayedlike micromorphologies compared to those with smooth micromorphologies. In conclusion, porous collagen scaffolds with various intrawall 3D micromorphologies can be constructed by application of dicarboxylic acids, superimposing the second level of morphology to the overall scaffold structure. Acid crystal formation is key to the specific micromorphologies observed and can be explained by the odd/even theory for dicarboxylic acids. Scaffolds with a 3D micrometer-defined topography may be used as a screening platform to select optimal substrates for the regeneration of specific tissues.
1. Introduction
The field of tissue engineering strives to reconstruct the structural and functional properties of damaged, diseased, or lost tissues.1 Its general strategy includes the construction of supportive biomaterials (scaffolds), providing a template guiding tissue formation, and the subsequent loading of scaffolds with biological compounds and cells.2 The selection of scaffolding materials is key since they have a considerable impact on the regenerative process.3 The interaction between cells and scaffolds is a complex interplay where scaffolds influence cell fate and cells modulate the scaffolds. Properties such as mechanical strength, degradation rate, pore size, and interconnectivity all influence cellular growth and function.4,5 Next to the overall morphology such as pore size and direction, micromorphologies in the scaffold may regulate cellular behavior. Three-dimensional (3D) topographical features have been shown to regulate cellular migration, attachment, viability, and differentiation.6,7 It has, for instance, been shown that bone marrow-derived mesenchymal stem cells preferentially adhere to ridged surfaces8 and that embryonic stem cells differentiate toward the neuronal lineage on ridged or grooved surfaces, without the use of any differentiation-inducing agents.9 Attachment and proliferation of chondrocytes are enhanced by the incorporation of ridged or grooved surfaces.10 Several physical and chemical micro- and nanofabrication patterning techniques are available to introduce specific morphologies (e.g., ridges, pillars, pits, and grooves) and distributions (e.g., random or regular). These techniques encompass soft lithography, photolithography, electrospinning, polymer phase separation, layer-by-layer microfluidic patterning, three-dimensional printing, chemical vapor deposition, ion milling, salt leaching, and reactive ion etching.11−13 The use of these techniques to introduce micromorphologies into scaffolds made from biological molecules is generally limited. In addition, the introduction of a specific micromorphology throughout the whole bioscaffold, rather than just to the surface, is challenging. We here show the application of dicarboxylic acids to introduce specific micromorphologies into the individual walls of porous collagen scaffolds and evaluate its effect on myoblasts cultured in vitro.
2. Experimental Section
2.1. Scaffold Construction
Type I collagen fibrils were isolated from bovine Achilles tendon as described. Collagen suspensions of 0.7% (w/v) were prepared using monocarboxylic and dicarboxylic acids. Monocarboxylic acids included formic acid (Merck, Darmstadt, Germany), acetic acid (Scharlau, Barcelona, Spain), and propionic acid (Merck). Dicarboxylic acids included oxalic acid (Merck), malonic acid (Sigma-Aldrich, St. Louis, MO), succinic acid (Sigma-Aldrich), glutaric acid (Sigma-Aldrich), maleic acid (Sigma-Aldrich), and fumaric acid (Sigma-Aldrich). An overview of the acids used (including their IUPAC names and chemical formula) and the molarities applied is provided in Table 1. After overnight incubation at 4 °C, collagen suspensions were homogenized on ice using a Teflon glass Potter–Elvehjem (Louwers Glass and Ceramic Technologies, Hapert, The Netherlands) with an intervening space of 0.35 mm (10 strokes) and deaerated by centrifugation at 117g for 30 min at 4 °C.14
Table 1. Overview of Acids and Concentrations Applied To Induce Micromorphologies in Unidirectional and Random Pore Collagen Scaffolds.
To investigate the effect of the various diluted acids on the micromorphology of collagen scaffolds, scaffolds with unidirectional and randomly oriented pores were constructed. Briefly, unidirectional collagen scaffolds were prepared by freezing collagen suspensions by directional freezing (10 mL/scaffold) using liquid nitrogen and a custom-made wedge system, followed by lyophilization in a Zirbus freeze dryer (Sublimator 500 II, Bad Grund, Germany).14 Random collagen scaffolds were prepared by freezing collagen suspensions in 6-well plates (5 mL/well) at −20 °C, followed by lyophilization.15 Scaffolds were strengthened by vapor fixation cross-linking using 37% formaldehyde (Scharlau) under vacuum for 30 min to improve the handling.14
2.2. Scanning Electron Microscopy
Scanning electron microscopy (SEM; JEOL SEM6340F, Tokyo, Japan) was used to investigate scaffold morphology. Scaffolds were cut longitudinally and perpendicularly, and samples were placed on stubs and sputtered with a thin layer of gold using a Polaron E5100 coating system.14 Images were recorded using an accelerating voltage of 10 kV. For pore size quantifications, images were recorded from four random locations of perpendicular samples. The lengths of the shortest axis of 50 pores were measured using ImageJ.14
2.3. Cell Culture
To evaluate the effect of micromorphologies on the differentiation of C2C12 murine skeletal muscle myoblasts, unidirectional collagen scaffolds with frayedlike (prepared using 0.25 M oxalic acid) and smooth (prepared using 0.25 M malonic acid) micromorphologies were selected.
Scaffolds were prepared as above and processed as described.16 Briefly, scaffolds were strengthened by vapor fixation with 37% formaldehyde (Scharlau) under vacuum for 30 min and processed into their final dimensions (diameter, 12 mm; height, 4 mm; top and bottom were removed) and remaining aldehydes were quenched by incubation in 1 M sodium phosphate buffer (pH 6.5, Merck, Darmstadt, Germany) containing 30 mM NaBH4 (Sigma-Aldrich, St. Louis, MO) for 1 h at 4 °C. Scaffolds were cross-linked using 33 mM N-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (Fluka Chemica AG, Buchs, Switzerland) and 6 mM N-hydroxysuccinimide (Fluka Chemica AG) in 50 mM 2-morpholinoethane sulfonic acid (MES buffer, pH 5.0; Sigma-Aldrich) containing 40% (v/v) ethanol for 4 h. Subsequently, scaffolds were washed with 0.1 M Na2HPO4 (Merck), 1 M NaCl (Merck), 2 M NaCl, demineralized water, and phosphate-buffered saline (PBS, pH 7.4) and sterilized while wet with 25 kGy γ-irradiation.16
Cell culture experiments were performed, n = 4, with each condition in triplicate. C2C12 murine skeletal muscle myoblasts were expanded in a monolayer until 90% confluency using proliferation medium: Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Carlsbad, CA), supplemented with 10% fetal calf serum (FCS, Pan-Biotech, Aidenbach, Germany), 100 IU/mL penicillin, and 100 μg/mL streptomycin (Amresco, Solon, OH). C2C12 cells were trypsinized using 0.05% trypsin–ethylenediamine tetraacetic acid (EDTA, Corning, Manassas, VA), followed by seeding of 2.5 × 106 cells per scaffold. In short, 100 μL of cell suspension (25 × 106 cells/mL) was dripped on the scaffolds while the scaffolds were placed on an autoclaved Whatman chromatography paper (3030-917, GE Healthcare Life Sciences, Pittsburg, PA) to allow the cell suspension to infiltrate the scaffold by capillary force. Scaffolds were then incubated at 37 °C for 3 h to let the cells adhere to the scaffolds, after which scaffolds were transferred to new 12-well plates and proliferation medium was added. The scaffolds were cultured for 7 days in proliferation medium and subsequently for 7 days in differentiation medium (DMEM), supplemented with 1% horse serum (PAA Laboratories, Cölbe, Germany), 100 IU/mL penicillin, and 100 μg/mL streptomycin. Cells cultured in the monolayer (5000 cells/well in 12-well plate) were used as a reference for quantitative polymerase chain reaction (qPCR) analysis. The medium was changed every 3 days. Scaffolds were harvested after 14 days and cut into half for histological qPCR analysis.
2.4. Histology and Immunohistochemistry
For histological evaluation, samples were washed in 0.1 M PBS, pH 7.4, fixed in 4% formaldehyde in 0.1 M PBS, pH 7.4, dehydrated through graded ethanol series, cleared in xylene, embedded in paraffin, and cut into 5 μm thick sections. Sections were stained with hematoxylin and eosin (H&E) or toluidine blue (2.5% in 5% Borax solution) and by immunofluorescence using an antibody against myosin.17 Briefly, sections were incubated in 0.1 M citrate buffer for 60 min, blocked by 0.15% glycine in 0.1 M PBS, pH 7.4, for 30 min, and in 1% (w/v) bovine serum albumin (BSA), 5% (v/v) normal goat serum, 0.1% (w/v) cold water fish gelatin (Sigma-Aldrich), and 0.1% (v/v) Triton X-100 in 0.1 M PBS, pH 7.4, for another 30 min. Sections were incubated with a primary antibody against myosin (1:400, MY-32, Sigma-Aldrich) for 60 min, followed by a secondary antibody goat antimouse IgG (H + L) Alexa 488 for 60 min and 10 μg/mL 4′,6-diamidino-2-phenylindole (DAPI; Roche Diagnostics GmbH, Mannheim, Germany) to stain nuclei. Control sections were incubated with the secondary antibody only. After each step, sections were washed with PBS. Sections were enclosed in Mowiol mounting medium (Sigma-Aldrich).
2.5. Quantitative PCR Analysis
Isolation of RNA, evaluation of RNA quality, synthesis of cDNA, and qPCR analysis were performed as described.16 Briefly, RNA was isolated in TRIzol (Life Technologies, Carlsbad, CA) using the RNeasy Mini kit (Qiagen GmbH, Hilden, Germany, 74106), followed by measuring RNA quality using a NanoDrop instrument (Thermo Scientific, Rockford, IL). An iScript cDNA synthesis kit (Bio-Rad Laboratories, Inc., Hercules, CA) was used to convert 500 ng of RNA into cDNA, using a T100 Thermal Cycler (Bio-Rad Laboratories, Inc.) for 5 min at 25 °C, 30 min at 42 °C, and 5 min at 85 °C. Milli-Q water was included as no template control (NTC). Obtained cDNA (20 μL) was diluted 20 times in Milli-Q water. Gene expressions were measured using SYBR Green qPCR, by iQ SYBR Green Supermix (Bio-Rad Laboratories, Inc.), and 2 μL of cDNA as a template, including NTC and Milli-Q water controls. The following primer sequences were used in real-time PCR analyses: glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward 5′-TGATGGGTGTGAACCACGAG-3′; reverse 5′-GGGCCATCCACAGTCTTCTG-3′; actinin forward 5′-TCATCCTCCGCTTCGCCATTC-3′; reverse 5′-CTTCAGCATCCAACATCTT-3′; and myosin heavy chain (pMHC) forward 5′-TCGCTGGGCTGGGTGTTAG-3′; reverse 5′-TGTCTGTCAGGCTGGGTGTG-3′. qPCR was performed on a CFX96 real-time system (Bio-Rad). The following amplification settings were used: 5 min at 95 °C, followed by 45 cycles of 10 s at 95 °C, 20 s at 60 °C, and 20 s at 72 °C. Crossing-point (Cp) values, expressed as quantification cycle (Cq), were obtained using Bio-Rad CFX Manager 3.1 software (Bio-Rad).16 Gene expressions were normalized to GAPDH and expressed as 2–ΔCq.
2.6. Statistics
Results of pore size measurements and qPCR are shown as mean ± standard deviation. Statistical differences were assessed using GraphPad Prism (GraphPad Software, Inc., version 5, La Jolla, CA) by unpaired t-tests and repeated measures analysis of variance (ANOVA) with Tukey’s posthoc tests to compare different groups, where p-values < 0.05 were considered statistically significant.
3. Results
3.1. Scaffold Characterization
3.1.1. Collagen Scaffolds Constructed with Dicarboxylic Acids
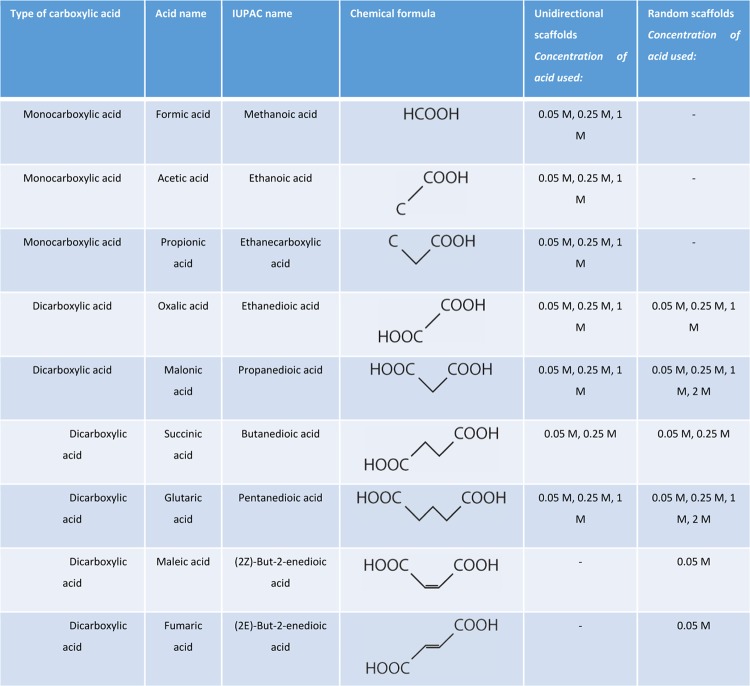
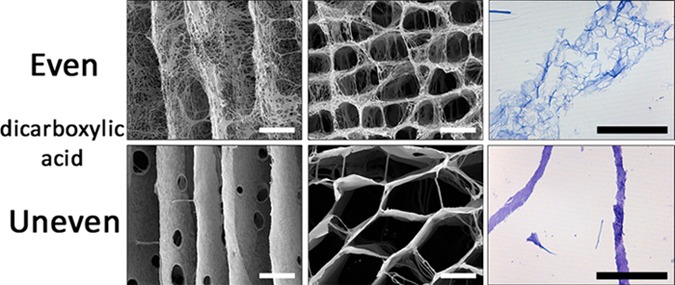
The construction of collagen scaffolds using various dicarboxylic acids resulted in striking different micromorphologies of the pore wall (Figure 1, unidirectional scaffolds). Morphology depended on whether acids were used with an even or odd (uneven) number of C-atoms. Oxalic acid (C2) and succinic acid (C4) showed morphologically open interconnective frayedlike wall structures, while the morphology in scaffolds constructed from malonic acid (C3) and glutaric acid (C5) showed smooth wall surfaces. Differences in frayedlike wall structures were observed: scaffolds prepared using oxalic acid displayed a honeycomb-like structure, whereas succinic acid resulted in a fibrillar micromorphology. Wall thickness also varied among the acids applied: oxalic acid and succinic acid resulted in thick walls, whereas the use of malonic acid and glutaric acid resulted in thinner walls.
Figure 1.
Morphology of scaffolds prepared using dicarboxylic acids. (A) Scanning electron micrographs of unidirectional scaffolds using different dicarboxylic acids (all 0.25 M) showing major variations in micromorphologies depending on the type of acid. The use of oxalic acid (C2) and succinic acid (C4) resulted in an open, frayedlike morphology, while malonic acid (C3) and glutaric acid (C5) resulted in smooth-walled structures. Specific frayedlike micromorphologies were observed: a honeycomb-like structure in walls from scaffolds prepared using oxalic acid, and a fibrillar structure using succinic acid. (B) Light microscopy images (toluidine blue staining) of unidirectional scaffolds prepared using oxalic acid or malonic acid. Note a frayed, open wall structure using oxalic acid, and a smooth, closed wall structure using malonic acid. (C) Scanning electron micrographs of longitudinal cross sections of unidirectional collagen scaffolds prepared using 0.05 and 1 M oxalic acid indicating that the pore morphology depends on the applied concentration, where a higher acid concentration increased wall thickness and frayed structure. Scale bars represent 50 μm.
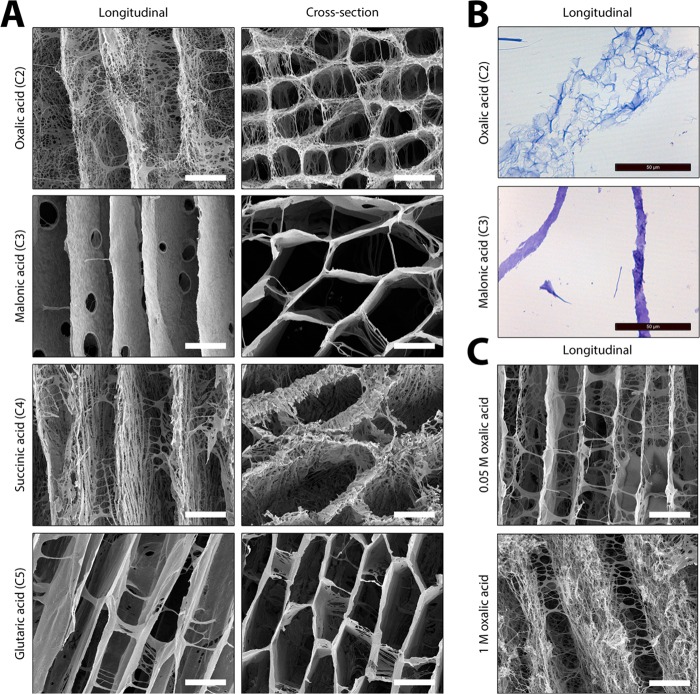
For collagen scaffolds with an overall random pore structure, a similar phenomenon was observed: a frayed, open wall structure when even-numbered acids (oxalic acid/succinic acid) were used (0.25 M), and a smooth, closed wall structure in the case of odd-numbered acids (malonic acid/glutaric acid; 0.25 M) (Figure 2). Morphologies were concentration-dependant. At very low concentrations (0.05 M), all acids resulted in a smooth appearance of the wall, whereas at high concentrations (2 M), the odd-numbered acids (malonic and glutaric acid) also resulted in frayed, open wall structures.
Figure 2.
Effect of type and concentration of dicarboxylic acid on the formation of micromorphologies in collagen scaffolds. Scanning electron micrographs of random pore scaffolds showing that scaffolds prepared using 0.05 M dicarboxylic acid have smooth wall structures. Pore wall structures are more frayed upon an increase in acid concentration. For dicarboxylic acids with an even number of C-atoms (oxalic acid and succinic acid), the pore structures are frayed at 0.25 M, while for odd acids (malonic acid and glutaric acid), these structures were observed at 1 M, being more apparent at 2 M. Scale bars represent 100 μm.
An increase in acid concentration generally resulted in more frayed open wall structures (Figure 2). For dicarboxylic acids with an even number of C-atoms, the pore structure became more frayed from 0.25 M acid onwards. For dicarboxylic acids with an odd number of C-atoms, frayed structures appeared when using 1 M acid, which became more apparent at concentrations of 2 M. These results indicate that the formation of smooth or frayed structures depends on the type as well as the concentration of the dicarboxylic acid (also see Section 3.1.3). An increase in acid concentration from 0.05 to 2 M reduced the pore size in scaffolds. For instance, in the case of oxalic acid, the pore size decreased from 143 ± 41 μm at 0.05 M to 89 ± 26 μm at 0.25 M (p < 0.0001); for glutaric acid, this was 132 ± 43 μm at 0.05 M to 36 ± 10 μm at 2.0 M (p < 0.0001).
Whereas there was a clear effect of the acids on the micromorphology of the wall structure of the individual pores, there was no effect on the overall structure of the scaffolds: the unidirectionality (Figure 1) or random pore structure (Figure 2) of scaffolds was not affected.
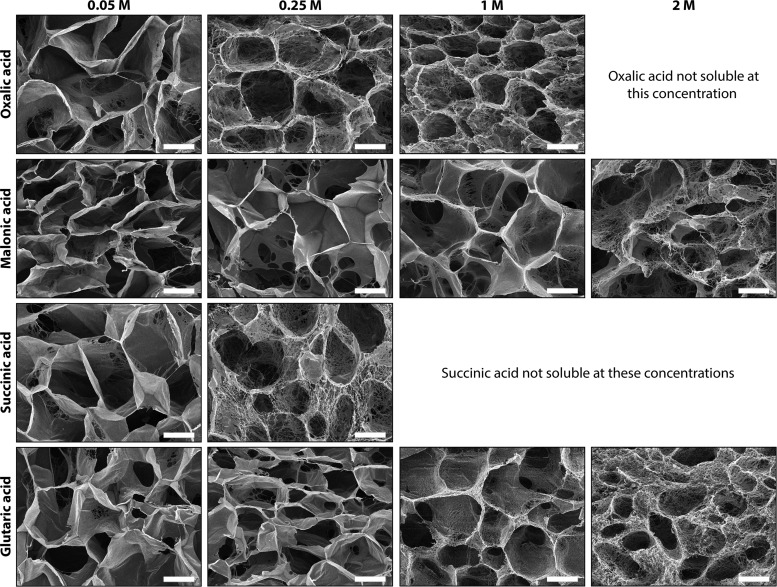
3.1.2. Collagen Scaffolds Constructed with Monocarboxylic Acids
No morphological differences in the pore wall structure and wall thickness were found between scaffolds prepared using the monocarboxylic acids, such as formic acid, acetic acid, and propionic acid (Figure 3 (unidirectional scaffolds); 0.25 M acid concentration: for SEM of random pore scaffolds, see refs (15, 18, 19) and (19)).
Figure 3.
Unidirectional scaffolds prepared using monocarboxylic acids. Scanning electron micrographs of unidirectional collagen scaffolds prepared using different monocarboxylic acids (all 0.25 M) showing no variations in micromorphologies. Scale bars represent 50 μm.
3.1.3. Effect of Unsaturated Dicarboxylic Acids with the Same Number of Carbon Atoms
To investigate the effect of acid solubility further, independently of the carbon chain length, random collagen scaffolds were constructed using the unsaturated C4 dicarboxylic acids, namely, maleic acid (well-dissolvable cis) and fumaric acid (poorly dissolvable trans). For maleic acid, smooth pore walls were visible, while a more threadlike morphology was observed for fumaric acid (Figure 4).
Figure 4.
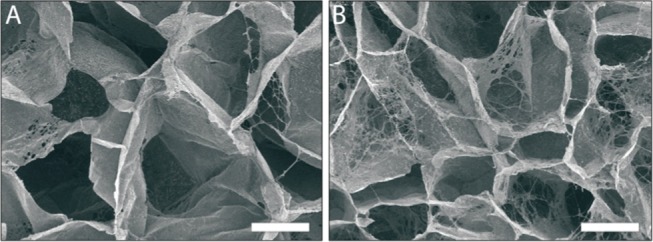
Effect of acid solubility on the formation of micromorphologies. Scanning electron micrographs of the pore structure of scaffolds prepared from cis/trans isomers (both C4), maleic acid (A, cis) and fumaric acid (B, trans), showed smooth and a more threadlike pore structure for maleic acid and fumaric acid, respectively. Scale bars represent 100 μm.
3.2. Cytocompatibility and Cellular Differentiation
3.2.1. General Morphology and Immunocytochemistry
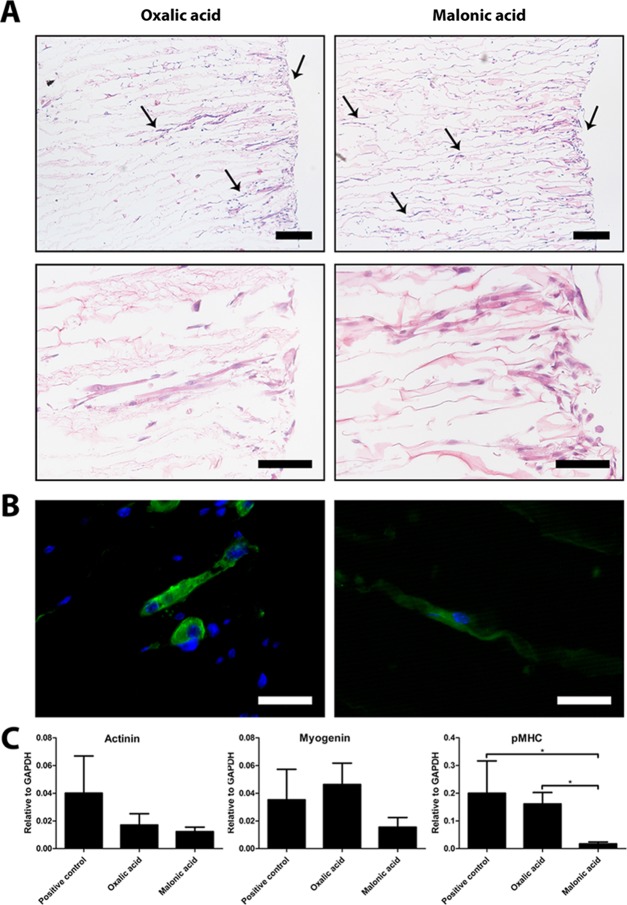
To evaluate the cytocompatibility of the scaffolds and to investigate the effect of incorporated micromorphologies on cell differentiation, C2C12 cells were cultured on unidirectional collagen scaffolds prepared using 0.25 M oxalic acid (frayedlike morphology) and 0.25 M malonic acid (smooth morphology) for 14 days. C2C12 cells infiltrated deeply into the scaffolds, which was facilitated by the unidirectional pore architecture. Using malonic acid, cells infiltrated deeper into the scaffolds compared to scaffolds prepared using oxalic acid (Figure 5A). Myosin staining (Figure 5B) was more intense for C2C12 cells cultured in scaffolds prepared using oxalic acid (frayed morphology).
Figure 5.
Differentiation of C2C12 cells seeded on scaffolds prepared using oxalic acid and malonic acid. (A) Unidirectional pore architecture facilitated infiltration of cells, but cells were found deeper in scaffolds prepared using malonic acid compared to those using oxalic acid (arrows represent the depth in the scaffold where cells were found). Scale bars represent 200 μm (top panel) and 100 μm (bottom panel). (B) More intense myosin staining for C2C12 cells cultured in scaffolds prepared from oxalic acid compared to those using malonic acid. Staining was more intense for oxalic acid. Scale bars represent 50 μm. (C) Analysis of gene expression levels of actinin (p = 0.081), myogenin (p = 0.064), and pMHC indicated that pMHC gene expression levels were significantly increased (p = 0.013) for C2C12 cells cultured on scaffolds prepared using oxalic acid (frayed morphology) compared to those using malonic acid (smooth morphology). A general trend was seen for higher gene expression levels in C2C12 cells cultured on frayed morphologies compared to that on smooth morphologies.
3.2.2. mRNA Analysis
Gene expressions of actinin, myogenin, and perinatal myosin heavy chain (pMHC) were analyzed in C2C12 cells and compared to the expression of the C2C12 cells cultured in the monolayer as a positive control. mRNA analysis indicated that pMHC gene expression was significantly upregulated for C2C12 cells cultured on scaffolds prepared using oxalic acid (frayed morphology) compared to those using malonic acid (smooth morphology). No differences were observed for actinin (p = 0.081) and myogenin (p = 0.064) gene expression levels between scaffolds prepared using oxalic acid and malonic acid and positive controls (Figure 5C). A general trend of higher gene expression levels of C2C12 cells was found when cultured on frayed morphologies compared to that on smooth morphologies.
4. Discussion
In vitro studies have investigated the effect of micromorphologies using two-dimensional (2D) monolayer culture models, where substrates have been modified with a number of topographical features, including ridges, pillars, pits, and grooves, applied in various patterns.11−13 These studies have been highly informative with regard to the 2D effect of specific micromorphologies on cellular behavior. The application of biological 3D patterned substrates like collagen scaffolds, however, has been limited. We here put forward a technology to incorporate specific 3D micromorphologies into individual walls of collagen scaffolds to study cellular behavior in a 3D setting. The technology is based on the use of specific dicarboxylic acids in the preparation of scaffolds.
In general, the overall pore structure of a scaffold is a reflection of the size and form of ice crystals formed during the freezing process. Upon lyophilization, the ice is sublimated, leaving behind a pore structure resembling the dimensions of the original ice crystal. In such a way, overall random and unidirectional pore structures can be introduced.14,15,20,21 In this study, we showed that the application of dicarboxylic acids adds the second level of morphology to the scaffolds, conferring a specific microstructure to the walls of individual pores. It was observed that α,ω-alkane dicarboxylic acids with an even number of C-atoms led to open, frayed/fibrillar wall structures, whereas dicarboxylic acids with an odd number of C-atoms led to scaffolds with a solid, smooth wall structure.
It is known that series of alkane derivatives often show alternating physicochemical characteristics between compounds with an even and odd number of carbon atoms, e.g., with respect to solubility.22 This odd–even effect is generally explained by a packing effect based on the shape of even- and odd-numbered alkanes resembling a parallelogram and a trapezoid, respectively,23,24 see Figure 6. Even-numbered compounds have their terminal groups on opposite sides of the zig-zag carbon chain, whereas in uneven-numbered compounds, the groups are on the same side. This makes uneven-numbered molecules relatively unsymmetric, fitting poorly into a crystal lattice. Therefore, the even-numbered ones fit better into a crystal lattice than the uneven-numbered ones. Dicarboxylic acids represent a specific group within the odd–even compounds. The formation of hydrogen-bonded dimers at both ends of the molecules (carboxyl dimer synthons) likely plays a role in this. Odd–even dicarboxylic acids differ in their capacity to form crystals. Thalladi et al.22 proposed a parallelogram–trapezoid model that explains the lower packing stability of odd acids by the repulsive effect of carboxy dimers of adjacent chains. In even acids, the distance between the carboxy dimers can be increased by slightly shifting each chain along the chain axis (lateral offset), thus decreasing repulsion. In odd acids, shifting the chain would increase the distance on one side of each molecule but decreases the distance on the other side, resulting in an increase in repulsion. The crystal packing of odd dicarboxylic acids therefore cannot be stabilized in this way, resulting in twisting of carboxy groups and severe torsion into molecules.22 Although this model is not directly translatable to the lower dicarboxylic acids (<C5), similar mechanisms may account for the solubility alternation found for these acids. Indeed, in a study on elastic moduli of dicarboxylic acids, Michra et al. confirmed the odd–even concept of Thalladi et al., including the C3 malonic acid and C4 succinic acid.25 It is however plausible that the influence of the hydrophobic effect on crystallization is less in the case of lower dicarboxylic acids.
Figure 6.
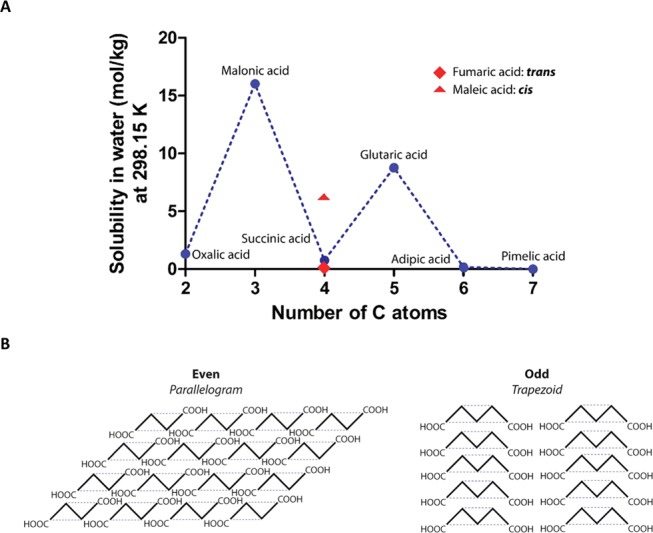
Relation between the number of C-atoms and dicarboxylic acid solubility and the crystal packing of even and odd acids. (A) Solubility of dicarboxylic acids in water. There is an even-odd effect with respect to the number of C-atoms on solubility. Differences in solubility are also present between fumaric acid and maleic acid (both 4 C-atoms), having low and high solubility, respectively. For poorly soluble acids, readily forming crystals and open, frayed/fibrillar structures within individual pore walls were observed in collagen scaffolds, whereas for highly soluble acids, smooth wall structures were found. (B) Overall structure of acids with an even and odd number of C-atoms (parallelogram versus trapezoid-like). The difficulty to form crystals in the case of trapezoid-shaped acids results in smooth pore walls, while the ease of crystallization of parallelogram-shaped acids results in frayed pore walls.
Taking into account the odd–even theory, the striking differences in the intrawall morphology observed for scaffolds prepared with various dicarboxylic acids may now be explained. During freezing of the scaffolds, the even dicarboxylic acids will crystallize and the crystals will be removed by lyophilization, leaving behind a micromorphology specific for the type of acid (compare the honeycomb structures for oxalic acid and the fibrillar structure for succinic acid, Figure 1). Odd-numbered acids will crystallize only at a very high (≥1 M) concentration. Thus, the ease or difficulty to form crystals from even (parallelogram) or odd (trapezoid) acids, respectively, give rise to the formation of micromorphologies observed in this study. The monocarboxylic acids tested (formic acid, acetic acid, and propionic acid) do not form specific microstructures, likely due to their high solubility in water, not readily forming crystals. Clearly, the solubility of the acid in water and the type of micromorphology are correlated. This was also observed for scaffolds constructed using the unsaturated dicarboxylic acids, maleic acid (cis isomer) and fumaric acid (trans isomer), both containing four carbon atoms. For the poorly soluble fumaric acid, a threadlike micromorphology was observed, whereas for the well-soluble maleic acid, a smooth morphology was noticed. The difference is solubility can be explained by the cis-conformation of maleic acids that enables the formation of intramolecular hydrogen bonds between the two carboxylic acid groups.26 This results in a strongly increased solubility (6.16 mol/kg at 298.15 K27) for maleic acid compared to that for fumaric acid (0.05–0.06 mol/kg).28
In vitro analysis was performed to investigate the cytocompatibility and the effect of different micromorphologies on cellular differentiation. C2C12 cells were cultured on unidirectional type I collagen scaffolds since these scaffolds facilitate cell infiltration due to the aligned pore architecture.14 In scaffolds with an open, frayedlike micromorphology, differentiation was favored in comparison to scaffolds with a smooth micromorphology, as indicated by increased myosin staining and pMHC gene expression level. This observation is in line with the preference of cells for irregular structures8−10 and may be explained by focal adhesion formation and subsequent signal transduction cascades. Yang et al.29 described that micromorphologies play important roles in the formation and dissociation of focal adhesions. Adaptation of focal adhesions due to the provided scaffold template, as a result of mechanotransduction, may result in physical changes and reorganization of intracellular compartments.30,31 Subsequently, this may result in differences in protein expression, interaction and concentration, and intracellular signaling, including that of focal adhesion kinase.32,33 The observation that cells in scaffolds with a smooth wall structure penetrate deeper into the scaffolds may be related to a lesser degree of interaction between cells and the substrate in comparison to frayed, rough wall structures.
In conclusion, this study shows that the specific intrawall micromorphology can be introduced to collagen scaffolds by the use of dicarboxylic acids. The micromorphologies correlate with the length of the carbon skeleton of the dicarboxylic acid and can be explained by the even/odd effect. Micromorphologies are adaptable by the type and concentration of the acids used and influence cell behavior.
Acknowledgments
We thank the Microscopic Imaging Centre for facilitating electron microscopy equipment (Radboud University Medical Center, Nijmegen, The Netherlands). This work was supported by a grant from the Dutch Government to the Netherlands Institute for Regenerative Medicine (NIRM, grant no. FES0908). The sources of funding have no other involvement in this publication.
Author Contributions
W.F.D. and T.H.v.K. contributed equally. The manuscript was written through the contribution of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
References
- Webber M. J.; Khan O. F.; Sydlik S. A.; Tang B. C.; Langer R. A perspective on the clinical translation of scaffolds for tissue engineering. Ann. Biomed. Eng. 2015, 43, 641–656. 10.1007/s10439-014-1104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer R.; Vacanti J. P. Tissue engineering. Science 1993, 260, 920–926. 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- Engler A. J.; Sen S.; Sweeney H. L.; Discher D. E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Keogh M. B.; O’Brien F. J.; Daly J. S. Substrate stiffness and contractile behaviour modulate the functional maturation of osteoblasts on a collagen-GAG scaffold. Acta Biomater. 2010, 6, 4305–4313. 10.1016/j.actbio.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Murphy W. L.; McDevitt T. C.; Engler A. J. Materials as stem cell regulators. Nat. Mater. 2014, 13, 547–557. 10.1038/nmat3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C. H.; Cheng Y. C.; Chao P. H. The influence and interactions of substrate thickness, organization and dimensionality on cell morphology and migration. Acta Biomater. 2013, 9, 5502–5510. 10.1016/j.actbio.2012.11.024. [DOI] [PubMed] [Google Scholar]
- Hulsman M.; Hulshof F.; Unadkat H.; Papenburg B. J.; Stamatialis D. F.; Truckenmuller R.; van Blitterswijk C.; de Boer J.; Reinders M. J. Analysis of high-throughput screening reveals the effect of surface topographies on cellular morphology. Acta Biomater. 2015, 15, 29–38. 10.1016/j.actbio.2014.12.019. [DOI] [PubMed] [Google Scholar]
- Tejeda-Montes E.; Smith K. H.; Poch M.; Lopez-Bosque M. J.; Martin L.; Alonso M.; Engel E.; Mata A. Engineering membrane scaffolds with both physical and biomolecular signaling. Acta Biomater. 2012, 8, 998–1009. 10.1016/j.actbio.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Lee M. R.; Kwon K. W.; Jung H.; Kim H. N.; Suh K. Y.; Kim K.; Kim K. S. Direct differentiation of human embryonic stem cells into selective neurons on nanoscale ridge/groove pattern arrays. Biomaterials 2010, 31, 4360–4366. 10.1016/j.biomaterials.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Park S. H.; Kim T. G.; Kim H. C.; Yang D. Y.; Park T. G. Development of dual scale scaffolds via direct polymer melt deposition and electrospinning for applications in tissue regeneration. Acta Biomater. 2008, 4, 1198–1207. 10.1016/j.actbio.2008.03.019. [DOI] [PubMed] [Google Scholar]
- Ghasemi-Mobarakeh L.; Prabhakaran M. P.; Tian L.; Shamirzaei-Jeshvaghani E.; Dehghani L.; Ramakrishna S. Structural properties of scaffolds: Crucial parameters towards stem cells differentiation. World J. Stem Cells 2015, 7, 728–744. 10.4252/wjsc.v7.i4.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloczko E.; Nikkhah D.; Yildirimer L. Scaffolds for hand tissue engineering: the importance of surface topography. J. Hand Surg. 2015, 10.1177/1753193415571308. [DOI] [PubMed] [Google Scholar]
- Griffin M. F.; Butler P. E.; Seifalian A. M.; Kalaskar D. M. Control of stem cell fate by engineering their micro and nanoenvironment. World J. Stem Cells 2015, 7, 37–50. 10.4252/wjsc.v7.i1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pot M.; Faraj K. A.; Adawy A.; van Enckevort W. J.; van Moerkerk H.; Vlieg E.; Daamen W. F.; van Kuppevelt T. H. A Versatile Wedge-Based System for the Construction of Unidirectional Collagen Scaffolds by Directional Freezing: Practical and Theoretical Considerations. ACS Appl. Mater. Interfaces 2015, 8495–8505. 10.1021/acsami.5b00169. [DOI] [PubMed] [Google Scholar]
- Faraj K. A.; van Kuppevelt T. H.; Daamen W. F. Construction of collagen scaffolds that mimic the three-dimensional architecture of specific tissues. Tissue Eng. 2007, 13, 2387–2394. 10.1089/ten.2006.0320. [DOI] [PubMed] [Google Scholar]
- Pot M. W.; de Kroon L. M. G.; van der Kraan P. M.; van Kuppevelt T. H.; Daamen W. F. Unidirectional BMP2-loaded collagen scaffolds induce chondrogenic differentiation. Biomed. Mater. 2018, 13, 015007 10.1088/1748-605X/aa8960. [DOI] [PubMed] [Google Scholar]
- Grefte S.; Adjobo-Hermans M. J.; Versteeg E. M.; Koopman W. J.; Daamen W. F. Impaired primary mouse myotube formation on crosslinked type I collagen films is enhanced by laminin and entactin. Acta Biomater. 2016, 30, 265–276. 10.1016/j.actbio.2015.11.009. [DOI] [PubMed] [Google Scholar]
- Pieper J. S.; Oosterhof A.; Dijkstra P. J.; Veerkamp J. H.; van Kuppevelt T. H. Preparation and characterization of porous crosslinked collagenous matrices containing bioavailable chondroitin sulphate. Biomaterials 1999, 20, 847–858. 10.1016/S0142-9612(98)00240-3. [DOI] [PubMed] [Google Scholar]
- Faraj K. A.; Brouwer K. M.; Geutjes P. J.; Versteeg E. M.; Wismans R. G.; Deprest J. A.; Chajra H.; Tiemessen D. M.; Feitz W. F. J.; Oosterwijk E.; Daamen W. F.; Kuppevelt T. H. The Effect of Ethylene Oxide Sterilisation, Beta Irradiation and Gamma Irradiation on Collagen Fibril-Based Scaffolds. Tissue Eng. Regener. Med. 2011, 8, 460–470. [Google Scholar]
- Schoof H.; Apel J.; Heschel I.; Rau G. Control of pore structure and size in freeze-dried collagen sponges. J. Biomed. Mater. Res. 2001, 58, 352–357. 10.1002/jbm.1028. [DOI] [PubMed] [Google Scholar]
- Hunger P. M.; Donius A. E.; Wegst U. G. Structure-property-processing correlations in freeze-cast composite scaffolds. Acta Biomater. 2013, 9, 6338–6348. 10.1016/j.actbio.2013.01.012. [DOI] [PubMed] [Google Scholar]
- Thalladi V. R.; Nusse M.; Boese R. The melting point alternation in alpha,omega-alkanedicarboxylic acids. J. Am. Chem. Soc. 2000, 122, 9227–9236. 10.1021/ja0011459. [DOI] [Google Scholar]
- Boese R.; Weiss H. C.; Blaser D. The melting point alternation in the short-chain n-alkanes: Single-crystal X-ray analyses of propane at 30 K and of n-butane to n-nonane at 90 K. Angew. Chem., Int. Ed. 1999, 38, 988–992. . [DOI] [PubMed] [Google Scholar]
- Bond A. D. On the crystal structures and melting point alternation of the n-alkyl carboxylic acids. New J. Chem. 2004, 28, 104–114. 10.1039/B307208H. [DOI] [Google Scholar]
- Mishra M. K.; Varughese S.; Ramamurty U.; Desiraju G. R. Odd-Even Effect in the Elastic Modulii of alpha,omega-Alkanedicarboxylic Acids. J. Am. Chem. Soc. 2013, 135, 8121–8124. 10.1021/ja402290h. [DOI] [PubMed] [Google Scholar]
- Leiserowitz L. Molecular Packing Modes - Carboxylic-Acids. Acta Crystallogr., Sect. B: Struct. Sci. 1976, 32, 775–802. 10.1107/S0567740876003968. [DOI] [Google Scholar]
- Apelblat A.; Manzurola E. Solubility of Oxalic, Malonic, Succinic, Adipic, Maleic, Malic, Citric, and Tartaric-Acids in Water from 278.15-K to 338.15-K. J. Chem. Thermodyn. 1987, 19, 317–320. 10.1016/0021-9614(87)90139-X. [DOI] [Google Scholar]
- Straathof A. J.; van Gulik W.. Production of Fumaric Acid by Fermentation. In Reprogramming Microbial Metabolic Pathways; Wang X.; C J.; Quinn P., Eds.; Springer: Netherlands, 2012; p 225. [DOI] [PubMed] [Google Scholar]
- Yang K.; Jung K.; Ko E.; Kim J.; Park K. I.; Kim J.; Cho S. W. Nanotopographical manipulation of focal adhesion formation for enhanced differentiation of human neural stem cells. ACS Appl. Mater. Interfaces 2013, 5, 10529–10540. 10.1021/am402156f. [DOI] [PubMed] [Google Scholar]
- McNamara L. E.; Burchmore R.; Riehle M. O.; Herzyk P.; Biggs M. J.; Wilkinson C. D.; Curtis A. S.; Dalby M. J. The role of microtopography in cellular mechanotransduction. Biomaterials 2012, 33, 2835–2847. 10.1016/j.biomaterials.2011.11.047. [DOI] [PubMed] [Google Scholar]
- Wang N.; Tytell J. D.; Ingber D. E. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 2009, 10, 75–82. 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- Lim S.-T. S. Nuclear FAK: a new mode of gene regulation from cellular adhesions. Mol. Cells 2013, 36, 1–6. 10.1007/s10059-013-0139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo B. K.; Wong S. T.; Lim C. K.; Kung T. Y.; Yap C. H.; Ramagopal Y.; Romer L. H.; Yim E. K. Nanotopography modulates mechanotransduction of stem cells and induces differentiation through focal adhesion kinase. ACS Nano 2013, 7, 4785–4798. 10.1021/nn304966z. [DOI] [PubMed] [Google Scholar]