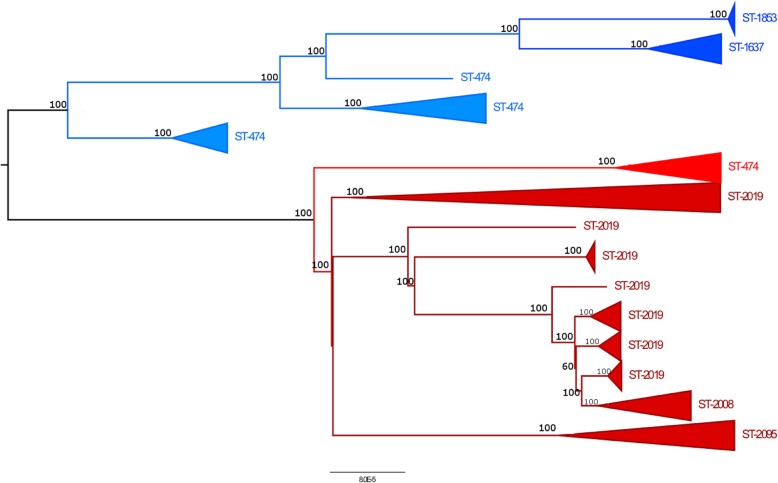
Fig. 2.
Core genome alignement-based ML phylogeny of S. Napoli genomes. Core genome alignment was used for phylogenetic reconstruction using RAxML (version 7.2.8, [27]) with bootstrapping and Maximum Likelihood (ML) search under the GAMMA model of rate heterogeneity. Tree visualization was obtained using FigTree v1.4.4 [28]. Subtrees were collapsed for ease of interpretation, while bootstrap values are indicated as node labels. Subtrees clearly group isolates belonging to the same ST; interestingly, two major clades can be identified (highlighted in blue and red, respectively), both containing isolates belonging to ST-474. This ST, consequently, appears to be biphyletic. Both subclades include also isolates belonging to different STs, other than ST-474, which are non-overlapping between the two subclades. Indeed, the first subclade groups ST-1637 and ST-1853 with ST-474; the second subclade groups ST-2019, ST-2008 and ST-2095 with ST-474