Figure 1.
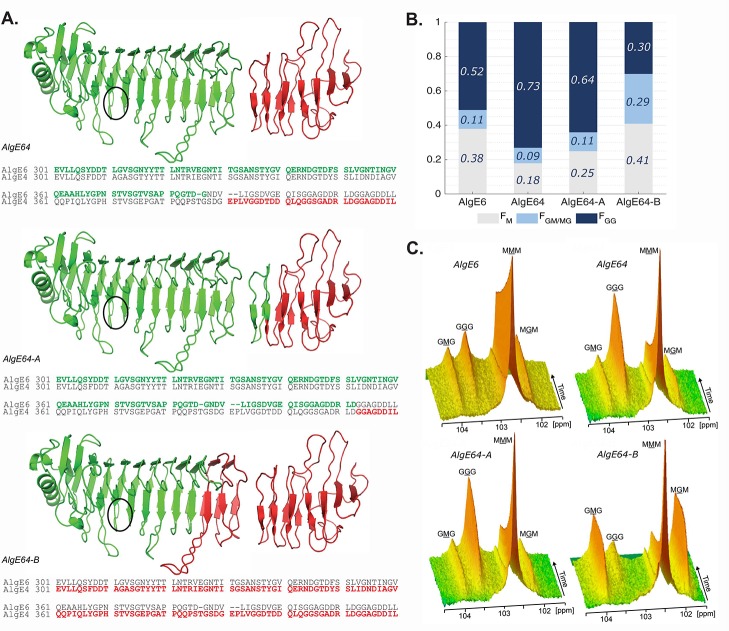
(A) Structural model of the hybrid enzymes AlgE64, AlgE64-A, and AlgE64-B, based on the crystal structure of AlgE4 A-module (PDB code 2PYG) and the NMR-structure of AlgE4 R-module (PDB code 2AGM). Parts in green belong to AlgE6, whereas red parts correspond to AlgE4. The location of the active site is indicated with black circles. The structures are visualized with PyMOL.46 Sequence alignment of the transition region between the A- and the R-modules in AlgE6 and AlgE4 is shown for each hybrid enzyme at the bottom of their ribbon structure. Residues colored in green (AlgE6) and red (AlgE4) denote the amino acids present in the corresponding hybrid epimerases. (B) Product composition at complete epimerization with AlgE6, and hybrid enzymes AlgE64, AlgE64-A, and AlgE64-B, calculated from 1H NMR spectra. M residues are shown in gray bars, GM/MG dyads in light blue bars, and GG-dyads in dark blue bars. On the y-axis is the fraction of each monad and dyad, whereas the four different enzymes are shown along the x-axis. These values are also shown in Table S1. (C) Time-resolved NMR spectra showing epimerization of 13C1-labeled poly-M for the four enzymes from B. The position of the triads in the spectra is indicated, and the M or G moiety generating the signal is underlined.