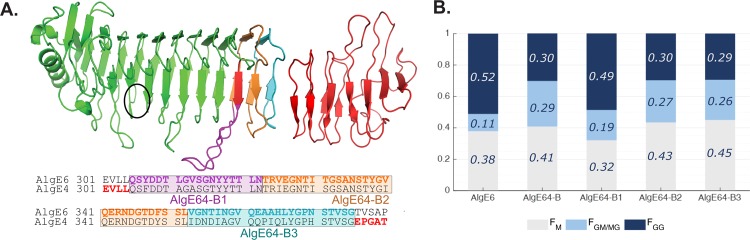
Figure 2.
(A) Structural model overview of AlgE64-B with localization of three regions (indicated by purple (AlgE64-B1), orange (AlgE64-B2), and, light blue (AlgE64-B3) colored parts) in the last part of the A-module, which were mutated to create three new mutants. In the three new mutants, amino acids belonging to AlgE4 are substituted to amino acids present in AlgE6. Sequence alignment of AlgE6 and AlgE4 is shown at the bottom of the structural model. Colored bold residues show the three different modified parts, the same color scheme as in the model. A sequence alignment of the long loop and how it differs between these three mutants and AlgE64-B is shown in Figure S2B. The location of the active site is indicated with a black circle. (B) Product composition at complete epimerization for AlgE6 and the four different AlgE64-B mutants, calculated from 1H NMR spectra. M residues are represented as gray bars, GM/MG dyads as light blue bars, and GG-dyads as dark blue bars. The y-axis denotes the fraction of the three product types, whereas the four different enzymes are listed on the x-axis. These values are also shown in Table S1.