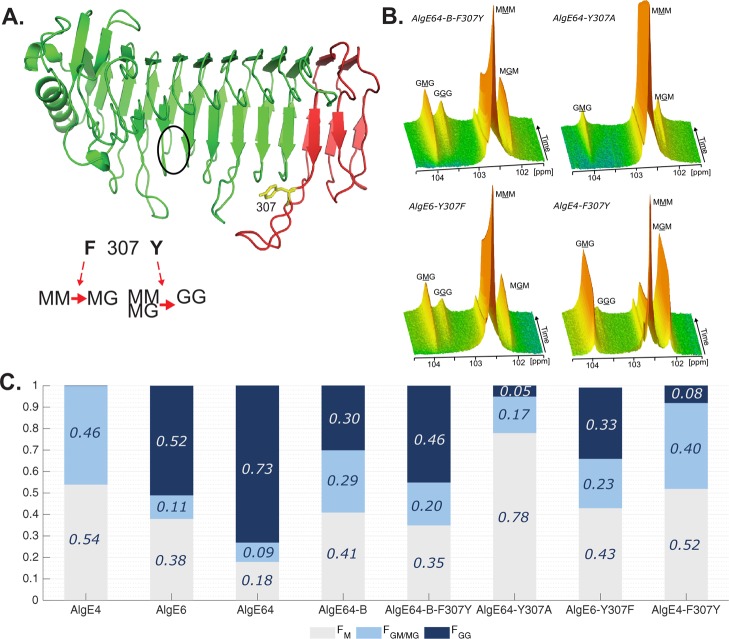
Figure 3.
(A) Ribbon structure of AlgE6 A-module represented in green (belonging to AlgE6) and red (belonging to AlgE4) colors as in point mutant AlgE64-B-F307Y. The location of the active site is indicated with a black circle. Tyrosine 307 in yellow is the point mutation of AlgE64-B-F307Y. Tyrosine 307 belongs to a loop in AlgE6 structure in proximity of the substrate binding groove. The model structure was obtained using SWISS-MODEL database.42 It is hypothesized that when residue 307 is a phenylalanine, the epimerases form an alternating (MG) block structure, whereas when it is a tyrosine they can form both MG and GG-blocks. (B) Time-resolved NMR spectra showing epimerization of 13C-labeled poly-M with AlgE64-B-F307Y, AlgE64-Y307A, AlgE6-Y307F, and AlgE4-F307Y. The position of the triads in the spectra is indicated, and the M or G moiety generating the signal is underlined. (C) Product composition at complete epimerization for the four enzymes from B and for AlgE4, AlgE6, AlgE64, and AlgE64-B, calculated from 1H NMR spectra. M residues are shown in gray bars, GM/MG dyads in light blue bars, and GG-dyads in dark blue bars. The y-axis denotes the fraction of each monad and dyad, whereas the seven different enzymes are listed on the x-axis. These epimerization patterns are also presented in Table S1.