Figure 1. Nuclear-Localized SPY, but Not SEC, Specifically Regulates the Circadian Clock.
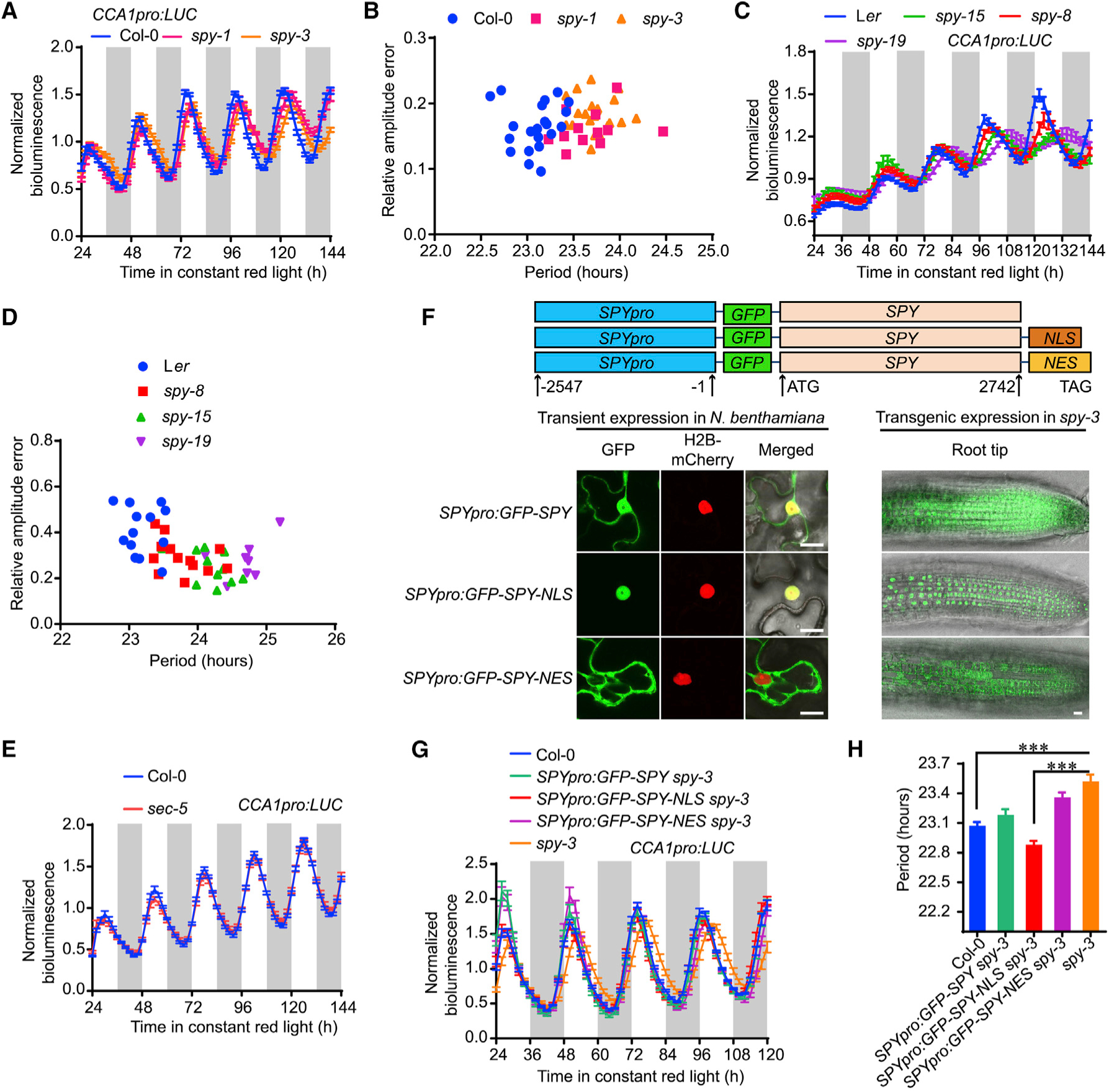
(A) Normalized bioluminescence activity of CCA1pro:LUC in spy-1, spy-3, and Col-0 plants in red light (20 μmol m−2 s−1). Data represent normalized means ± SE (n = 13–21). White and light-gray vertical bars indicate the subjective day and night, respectively.
(B) Scatterplot showing lengthened circadian periods in spy mutants with relatively normal amplitude errors, indicating strong circadian robustness in the estimation of circadian period.
(C) Normalized bioluminescence activity of CCA1:LUC in spy mutants with Ler background. Data represent means ± SE after normalization with the average bioluminescence intensity over 24 h to 144 h.
(D) Scatterplot showing the lengthening circadian period in spy mutants, with a relative amplitude error lower than 0.5.
(E) Null mutant of sec did not show aberrant circadian period phenotype, as indicated by bioluminescence trace of CCA1pro:LUC (n = 20).
(F) Subcellular localization of SPY with or without nuclear localization signal (NLS) and nuclear export signal (NES) in both the epidermal cells of transiently infiltrated N. benthamiana leaves and the root tips of stable transgenic Arabidopsis. Histone H2B-mCherry was used as a nuclear marker. Scale bars, 10 μm.
(G) Normalized bioluminescence activity of CCA1pro:LUC in SPY:GFP-SPY spy-3 and SPY:GFP-SPY-NLS spy-3. Data represent means ± SE (n = 18).
(H) The estimated circadian periods from Figure 1G showing that SPY:GFP-SPY-NLS could fully rescue the lengthened circadian period in spy-3 mutant. Data represent means ± SE (n = 18). *** indicates significant difference at p < 0.001 in t-test.
