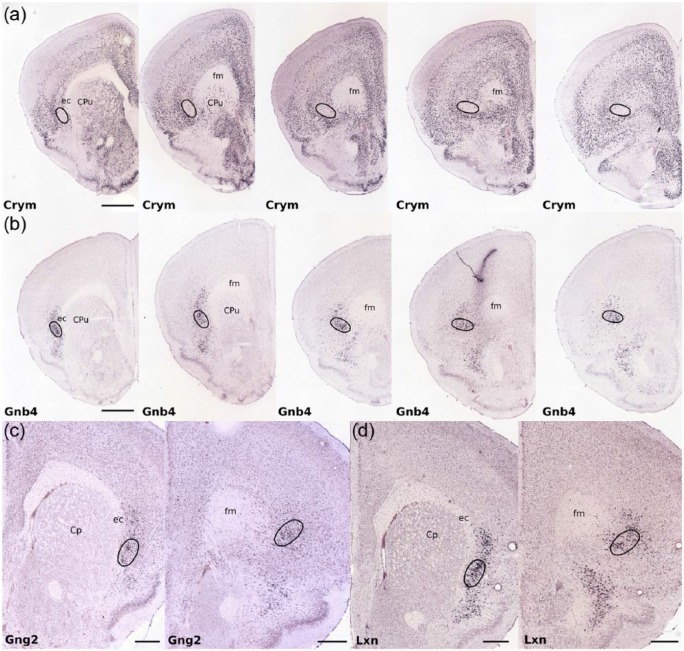
Figure 1.
Photomicrographs showing the nucleotide sequence expression (in situ hybridisation) of four example genes that are enriched or attenuated in the claustrum as identified by Wang et al. (2017). In each example claustrum-specific label extends rostral to the anterior apex of the striatum. (a) Crystallin mu (Crym) expression is attenuated in the claustrum but enriched in surrounding cortex delineating a claustral boundary that extends anterior to the claustrum, maintaining an ovoid cross-section at the ventro-lateral apex of the forceps minor of the corpus callosum (fm). (b) Gnb4 (Guanine nucleotide binding protein (G protein), beta 4) expression is enriched in the claustrum throughout the claustrum (Wang et al., 2017), including its rostral extent as defined by the Cyrm template in (a). (c) Gng2 (guanine nucleotide binding protein (G protein), gamma 2) expression is enriched in the claustrum (Mathur et al., 2009), again extending rostral to the anterior apex of the striatum. (d) Lxn (Latexin) expression is also enriched throughout the claustrum including its rostral extent. Of the remaining 45 genes identified by Wang et al. (2017), 44 showed equivalent differential expression in the rostral claustrum when compared to the ‘claustrum proper’. Black oval outlines delineate the claustral border in each case. The images were taken from the Allen Institute for Brain Science Mouse Brain Atlas, Allen Institute for Brain Science, Allen Mouse Brain Atlas. Available at: http://mouse.brain-map.org/