Abstract
TANK Binding Kinase 1 (TBK1) is an important kinase involved in innate immune response. Here we discover that TBK1 is hyperactivated by von Hippel-Lindau (VHL) loss or hypoxia in cancer cells. Tumors from kidney cancer patients with VHL loss display elevated TBK1 phosphorylation. Loss of TBK1 via genetic ablation, pharmacological inhibition, or a new Cereblon-based proteolysis targeting chimera (PROTAC) specifically inhibits VHL-deficient kidney cancer cell growth, while leaving VHL wild type cells intact. TBK1 depletion also significantly blunts kidney tumorigenesis in an orthotopic xenograft model in vivo. Mechanistically, TBK1 hydroxylation on Proline 48 triggers VHL as well as the phosphatase PPM1B binding that leads to decreased TBK1 phosphorylation. We identify that TBK1 phosphorylates p62/SQSTM1 on Ser366 which is essential for p62 stability and kidney cancer cell proliferation. Our results establish that TBK1, distinctive from its role in innate immune signaling, is a synthetic lethal target in cancer with VHL loss.
Keywords: VHL, TBK1, hydroxylation, ccRCC, EglN1
INTRODUCTION
Estimated new cases and deaths from renal (renal cell and renal pelvis) cancer in the U.S. in 2014 were 63,920 and 13,860 respectively(1). Kidney cancer incidence has been increasing steadily for the past several decades, although the reasons for this are unclear(1). The VHL tumor suppressor gene was identified as a germline mutation in patients at risk for ccRCC, which accounts for approximately 85% of all kidney cancers(2). Inactivating VHL mutations also play major roles in sporadic kidney cell cancer(3). It is well established that the pVHL-associated complex has E3 ubiquitin ligase activity and VHL loss leads to hypoxia inducible factor α (HIFα, including HIF1α and HIF2α) and ZHX2 stabilization, which contributes substantially to the transforming phenotype of renal cancer (2,4–7). Further research show that pVHL interacts with HIF1α through the hydroxylation of defined HIF1α proline residues (prolines 402 and 564) by members of the EglN family of iron- and 2-oxoglutarate-depedent dioxygenases (EglN1, EglN2, and EglN3)(8). As a result of accumulation and translocation of HIFα factors into the nucleus, HIFαs dimerize with a constitutively expressed HIFβ-subunit and transactivate genes that have hypoxia response elements (NCGTG) in promoters or enhancer regions, such as genes involved in angiogenesis (e.g. VEGF), glycolysis and glucose transport (e.g. GLUT1) and erythropoiesis (e.g. EPO)(9). HIF signaling/activation is an important oncogenic signature for VHL-deficient ccRCC. However, it remains challenging to target HIF signaling in ccRCC. HIF2α stabilization, as a result of pVHL loss, is sufficient and necessary for promoting kidney tumor growth (7). Recent reports showed that the specific HIF2α inhibitor PT2399 inhibits primary tumor growth and invasion of a subset of kidney cancer (10,11). However, a significant portion of kidney cancer remains resistant to HIF2α inhibitor treatment (10,11), highlighting the importance of identifying additional therapeutic vulnerabilities of VHL-deficient kidney cancer.
Tumor specific genetic alteration (such as VHL loss) reveals not only the biological changes that drive tumor progression but also vulnerabilities that can be exploited therapeutically. Since 70–80% of kidney tumors harbor VHL functional loss, it remains very attractive to identify synthetic lethality partners for VHL loss in kidney cancer while sparing normal cells. Previous research has identified a handful of pharmacological inhibitors, including autophagy modulator STF-62247(12), homoharringtonine (HHT) (13), EZH inhibitors (14), GLUT-1 inhibitors (15) and ROCK inhibitors (16), displayed the selective killing of VHL null ccRCC cells. In addition, CDK6, MET and MAP2K1 were reported to be essential for ccRCC cell lines with VHL loss (17). Some of these pathways are known HIF signaling regulators while the mechanisms for other VHL synthetic lethality partners remains unknown.
TBK1 is a member of the atypical IκB kinase (IKK) family, which also features another highly related family member IKKε. Upon DNA and RNA virus infection, Stimulator of Interferon Gene (STING) binds with TBK1 and promotes its phosphorylation on Ser172 within the TBK1 activation loop, which is necessary for its kinase activity to induce STING phosphorylation on Ser366 and the type I interferon response by directing IRF3 phosphorylation (18,19). As such, TBK1 is a required element of innate immune signaling in cells. In recent years, the role of TBK1 has been expanded into cancers (20,21). Although previous research suggested that RalB/Sec5 effector or Axl signaling may act upstream of TBK1 signaling (22,23), it is largely unclear on how TBK1 activity is dynamically regulated in cancers and whether this activation is connected to its canonical signaling in innate immunity. Here we identify a novel role of TBK1 signaling in cancer, distinctive from its role in innate immune signaling, by serving as a synthetic lethal partner for VHL null kidney cancer in a HIF independent manner.
RESULTS
VHL Suppresses TBK1 Activity in ccRCC
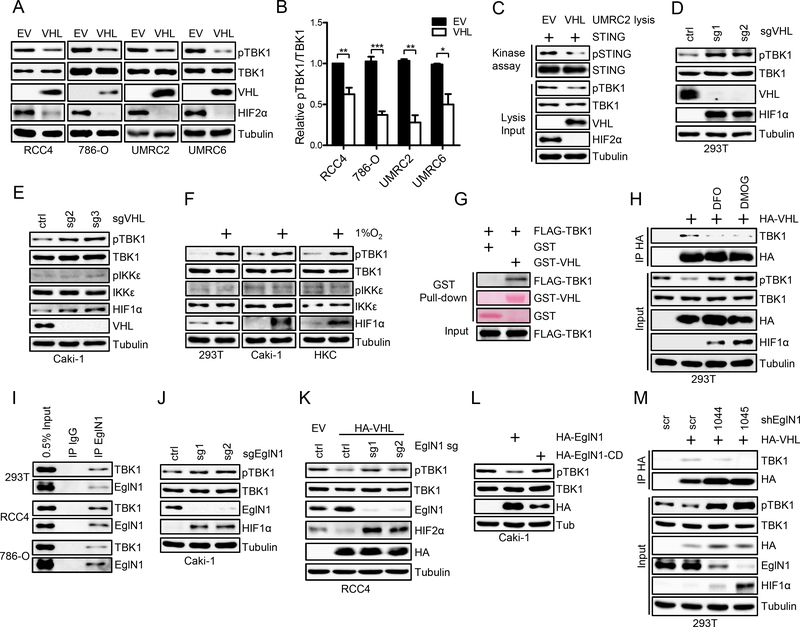
By using a pan-prolyl hydroxylation antibody to perform pull down followed by mass spectrometry analysis in Hela cell lysates, TBK1 was indicated to be hydroxylated (24). Since cells were not treated by MG132, many VHL degradation substrates may not be retrieved from the pull down, including HIF1α and HIF2α (24). Among the list that were pulled down from the mass spectrometry, TBK1 is one of the handful kinases that may be therapeutically targetable. Because hydroxylated protein may interact with and be potentially regulated by VHL, we set to determine whether TBK1 protein level or its canonical phosphorylation on Ser172, that governs its activity, may be regulated by VHL. To this end, we examined TBK1 or p-TBK1 (Ser172) levels in ccRCC isogenic cell lines (786-O, UMRC2, RCC4 and UMRC6) that are either VHL null (with empty vector (EV)) or with VHL restoration. Interestingly, whereas total TBK1 protein levels did not change upon VHL expression, phosphorylated TBK1 was significantly suppressed by VHL in all four cell lines examined (Fig. 1A and B). Similar regulation was also found in 293T cells overexpressing VHL (Supplementary Fig. S1A). To show whether the level of pTBK1 is a good predictor for TBK1 kinase activity, we performed in vitro kinase assay using UMRC2 cell lysates as the source of the kinase and purified STING protein as a substrate. STING phosphorylation was detected by a STING phospho-Ser366 antibody. As a control, we depleted TBK1 in UMRC2 cells by specific sgRNA and found that TBK1 depletion led to decreased STING phosphorylation on Ser366 (Supplementary Fig. S1B), suggesting that this kinase assay can be used to reliably detect TBK1 activity. We found that lysates from UMRC2 empty vector cells displayed stronger STING phosphorylation than lysates from the isogenic cells restored with VHL (Fig. 1C). Conversely, we also depleted VHL by several independent sgRNAs with CRISPR-Cas9 in VHL proficient cells (293T and Caki-1). Consistently, VHL depletion led to profound upregulation of p-TBK1 while not affecting total TBK1 levels (Fig. 1D and E). It is worth mentioning that the protein levels and phosphorylation state of IKKε, a close family member of TBK1, was not affected by VHL in these cells (Fig. 1E; Supplementary Fig. S1C), suggesting that VHL regulation of TBK1 phosphorylation is specific. IL-1 β and TNF-α can activate TBK1 activity independent of adaptors such as STING (25). To exclude the potential effect of these cytokines on TBK1 signaling, we examined the conditional medium from EV and isogenic VHL cell culture. While TNF-α in most medium sample is lower than detection limit, IL-1β level is comparable in EV and VHL medium (Supplementary Fig. S1D).
Figure 1. VHL/EglN1 interacts with TBK1 and suppresses TBK1 activity.
A, Immunoblots of lysates from RCC4, 786-O, UMRC2 and UMRC6 cells restored with empty vector (EV) or VHL.
B, Quantification of pTBK1/TBK1 signal from panel A performed by Image J. n=3 biological replicates.
C, Immunoblots of samples of kinase assay using UMRC2 lysates as kinase and STING protein as substrate.
D-E, Immunoblots of lysates from 293T and Caki-1 cells infected with lentivirus encoding either Ctrl sgRNA or VHL sgRNA.
F, Immunoblots of lysates from 293T, Caki-1 and HKC cells without or with hypoxia (1% O2, overnight) treatment.
G, Immunoblots or ponceau S staining of samples from pull-down binding assay with purified GST, GST-VHL and in vitro translated FLAG-TBK1.
H, Immunoblots of whole cell lysates (input) and immunoprecipitations (IP) from 293T cells transfected with vector or HA-VHL and then treated with DMOG or DFO as indicated.
I, Immunoblots of whole cell lysates (input) and immunoprecipitations (IP) from 293T, RCC4 and 786-O cells.
J-L, Immunoblots of lysates from Caki-1, RCC4EV and RCC4VHL cells infected with lentivirus encoding either Ctrl or EglN1 sgRNA or transfected with indicated plasmids.
M, Immunoblots of whole cell lysates (input) and immunoprecipitations (IP) from 293T cells infected with lentivirus encoding scramble shRNA (scr) or EglN1 shRNA sequences and then transfected with vector or HA-VHL as indicated.
VHL loss creates a pseudo-hypoxia condition in ccRCC that is characterized with constitutive HIF stabilization (26). Hypoxia is also a characteristic of most solid cancers (27). We exposed VHL proficient cells (293T, Caki-1 and HKC) and RCC4 cells expressing exogenous HA-VHL with hypoxia (1% O2) and also found consistent p-TBK1 upregulation (Fig. 1F; Supplementary Fig. S1E). As an orthogonal approach, we treated VHL proficient Caki-1 cells with hypoxia mimetics (DMOG, DFO) or VHL-restored RCC4, 786-O or UMRC2 cells with FG4592 or DFO. In all cases, we found TBK1 phosphorylation was significantly upregulated (Supplementary Fig. S1F and S1G). To address the question of whether this upregulation was due to HIF upregulation, we depleted HIF2α expression by two independent sgRNAs and found that TBK1 phosphorylation level was not decreased by HIF2α depletion (Supplementary Fig. S1H). We also used two independent sgRNAs to deplete ARNT, an essential binding partner of HIF2α for its transcriptional activity, in 786-O and UMRC2 cells. Consistently, we did not find that TBK1 phosphorylation was affected by ARNT depletion (Supplementary Fig. S1I). Altogether, our results suggest that VHL or hypoxia regulates TBK1 phosphorylation in a HIF-independent fashion.
To further determine whether VHL-mediated p-TBK1 downregulation is mediated by prolyl hydroxylation, we treated HA-VHL-restored 786-O cells with prolyl hydroxylase inhibitors (DMOG, DFO) and found that these inhibitors upregulated p-TBK1 level in VHL restored cells to a similar level as in the vector cells (Supplementary Fig. S1J). To examine whether TBK1 and VHL directly interact, we performed GST-VHL pull down with in vitro translated FLAG-TBK1 and found a direct interaction between them (Fig. 1G). To determine whether the interaction of VHL and TBK1 mediates TBK1 phosphorylation regulation by VHL, we examined their binding in the absence or presence of prolyl hydroxylase inhibitors (DFO and DMOG) and found that prolyl hydroxylase inhibitors eliminated the binding between TBK1 and VHL, which corresponded with increased TBK1 phosphorylation (Fig. 1H). Consistently, DFO and DMOG treatment also led to increased TBK1 phosphorylation and activity in VHL restored UMRC2 cells (Supplementary Fig. S1K). Therefore, our data suggest that prolyl hydroxylation may promote the interaction between TBK1 and VHL therefore contributing to decreased TBK1 phosphorylation and activity.
EglN1 Prolyl Hydroxylase Regulates TBK1-VHL interaction and TBK1 phosphorylation
To address the prolyl hydroxylase that may contribute to TBK1 hydroxylation and its regulation by VHL, we examined the interaction between TBK1 and three EglN family members (EglN1, 2 and 3) and found that EglN1 is the primary prolyl hydroxylase that showed the most robust binding with TBK1 (Supplementary Fig. S2A). Additionally, we showed that EglN1 and TBK1 could bind endogenously in several cell lines including 293T, RCC4 and 786-O (Fig. 1I). Next, we depleted EglN1 by two independent sgRNAs and found that EglN1 depletion led to increased TBK1 phosphorylation (Fig. 1J). In addition, depletion of EglN1 in HA-VHL expressing RCC4 cells upregulated TBK1 phosphorylation to a similar level as in the RCC4 cells with vector control (Fig. 1K). As an orthogonal approach, we also showed that TBK1 phosphorylation was upregulated in EglN1 knockout MEFs, but not in EglN2/3 knockout MEFs (Supplementary Fig. S2B). Additionally, we also depleted EglN1 with two independent shRNAs in 293T cells and consistently observed increased TBK1 phosphorylation (Supplementary Fig. S2C). In these cells with EglN1 depletion, DMOG treatment could not further upregulate TBK1 phosphorylation, suggesting that EglN1 is the major hydroxylase enzyme that regulates TBK1 phosphorylation (Supplementary Fig. S2C). Conversely, we also overexpressed EglN1 in Caki-1 cells and observed decreased TBK1 phosphorylation with wild type but not a catalytically dead EglN1 mutant (EglN1-CD, H374A/D376A) (Fig.1L). This regulation is further confirmed by the upregulation of TBK1 phosphorylation in EglN1 overexpressing cells treated with prolyl hydroxylase inhibitors FG4592 or DFO (Supplementary Fig. S2D and S2E). To examine whether EglN1 regulates the interaction between TBK1 and VHL therefore contributing to TBK1 phosphorylation regulation, firstly we performed in vitro hydroxylation followed by VHL binding assay. In the presence of EglN1, the binding between TBK1 and VHL increased, suggesting that EglN1-mediated hydroxylation on TBK1 promotes TBK1-VHL interaction (Supplementary Fig. S2F). We then depleted EglN1 by two independent shRNAs and found that EglN1 depletion led to decreased binding between TBK1 and VHL, which corresponded with increased TBK1 phosphorylation (Fig. 1M). Overall, our results suggest that EglN1 hydroxylates TBK1 which leads to its binding with VHL and decreased TBK1 phosphorylation.
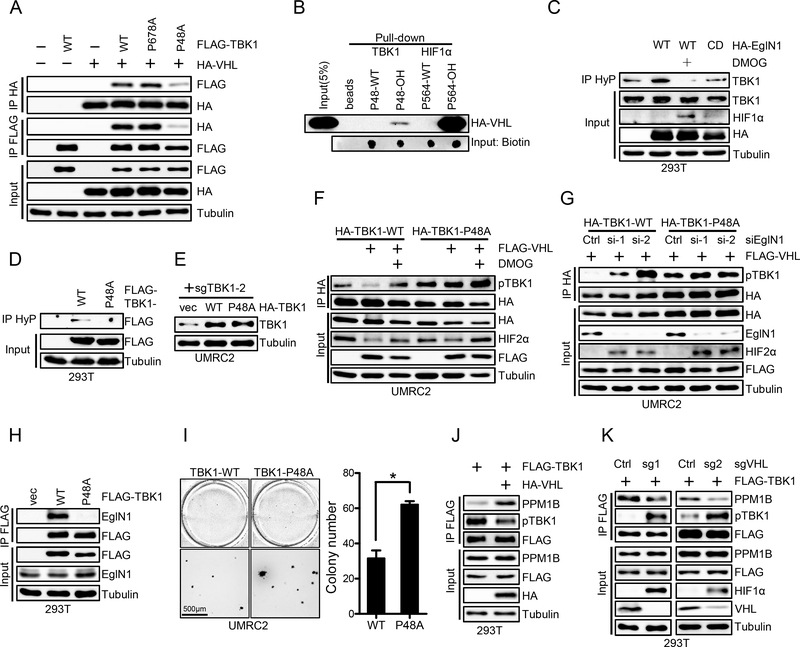
Next, we aimed to identify TBK1 prolyl hydroxylation sites that may be important for its regulation by EglN1 and VHL. To this end, we overexpressed FLAG tagged TBK1 in 293T cells followed by either DMSO or DMOG treatment and analyzed immunoprecipitated FLAG-TBK1 via mass spectrometry to identify potential hydroxylation sites. We were specifically interested in potential proline sites that were hydroxylated, but whose hydroxylation level were diminished upon DMOG treatment. We identified two such prolyl hydroxylation sites: Proline 678 and 48 (Supplementary Fig. S3A and S3B). Next, we mutated TBK1 Proline 678 and 48 to Alanines (P678A, P48A). Whereas TBK1 WT or P678A mutant binds with VHL efficiently, P48A mutant displayed diminished binding with VHL confirmed by reciprocal immunoprecipitations (Fig. 2A), suggesting that Proline 48 is the major hydroxylation site that binds with VHL. Subsequently, we synthesized TBK1 WT or P48-OH peptides and performed binding assay with VHL. Among these peptides, only Proline 48 hydroxylated peptide binds with VHL, although considerably weaker compared with the binding of hydroxylated HIF1α peptide with VHL (Fig. 2B). We also performed immunoprecipitation using a pan-hydroxyl proline antibody and found that WT, but not catalytic dead EglN1 or EglN1 in combination with DMOG, promoted TBK1 hydroxylation (Fig. 2C). Consistent with the notion that proline 48 is the major TBK1 hydroxylation site, pan-hydroxylation IP followed by FLAG immunoblot also confirmed that P48A mutant abrogated all hydroxylation signals present in WT TBK1 (Fig. 2D). To determine whether proline 48 site is the major site affecting TBK1 phosphorylation, we depleted endogenous TBK1 from UMRC2 cells and re-introduced either WT or P48A TBK1 into these cells (Fig. 2E). In WT TBK1 expressing cells, VHL overexpression led to decreased TBK1 phosphorylation that can be upregulated upon concurrent treatment with the prolyl hydroxylase inhibitor DMOG (Fig. 2F). On the other hand, cells expressing TBK1 P48A mutant displayed constitutive TBK1 phosphorylation that is resistant towards VHL expression (Fig. 2F). It is important to note that TBK1 P48A mutant also showed higher TBK1 phosphorylation at the basal level despite a lower total TBK1 level. Next, we also wanted to examine whether TBK1 hydroxylation by EglN1 is mainly contributing to the phosphorylation regulation, and we found that TBK1 phosphorylation was upregulated by EglN1 depletion by two independent EglN1 siRNAs in wild type TBK1 expressing cells (Fig. 2G). On the other hand, TBK1 P48A mutant displayed constitutive upregulation of phosphorylation that was resistant to EglN1 depletion (Fig. 2G). According to crystal structure of TBK1 dimer, P48 residue sits on the periphery of TBK1 kinase domain that might be accessible by EglN1 (Supplementary Fig. S3C). We then examined the binding between EglN1 and TBK1-WT or P48A mutant and found that P48A could not bind with EglN1, suggesting that Proline 48 site is the major EglN1 hydroxylation site (Fig. 2H). Finally, VHL restored UMRC2 cells harboring TBK1-P48A mutant could form more colonies compared to cells harboring TBK1 WT in 3-D soft agar growth assay (Fig. 2I). Although IKKε and TBK1 exhibits highly similarity, the sequences surrounding Pro48 are distinct in these two proteins (Supplementary Fig. S3D), suggesting that Pro48 mediated regulation is specific for TBK1. Furthermore, TBK1 Pro48 (LRP motif) is highly conserved in all vertebrates, suggesting its functional importance (Supplementary Fig. S3E). In summary, TBK1 is hydroxylated on Pro48 residue which promotes VHL binding and its decreased phosphorylation.
Figure 2. Hydroxylated Pro48 mediates TBK1-VHL interaction.
A, Immunoblots of whole cell lysates (input) and immunoprecipitations (IP) from 293T cells transfected with indicated plasmids.
B, Immunoblots of peptide pull-down samples as indicated.
C-D, Immunoblots of whole cell lysates (input) and immunoprecipitations (IP) from 293T cells transfected with indicated plasmid and treated with DMOG (1mM) as indicated. HyP denotes pan-hydroxyPro antibody.
E, Immunoblots of lysates from UMRC2 cells infected with lentivirus encoding vec, HA-TBK1 or HA-TBK1-P48A and then infected with lentivirus encoding TBK1 sgRNA(sg2).
F-G, Immunoblots of whole cell lysates (input) and immunoprecipitations (IP) from UMRC2 cells infected with lentivirus encoding HA-TBK1-WT or HA-TBK1-P48A and transfected with indicated plasmids or siRNA, followed by indicated treatment.
H, Immunoblots of whole cell lysates (input) and immunoprecipitations (IP) from 293T cells transfected with indicated plasmids.
I, Representative soft agar growth and quantification of colony number (duplicate wells) of UMRC2 cells infected with lentivirus encoding HA-TBK1 or HA-TBK1-P48A and then transfected with HA-VHL. See Fig. 2E for HA-TBK1 expression. Error bars represent SEM, * P<0.05.
J-K, Immunoblots of whole cell lysates (Input) and immunoprecipitations (IP) from 293T cells infected with indicated lentivirus and transfected with indicated plasmids.
The next question was how VHL binding to TBK1 may contribute to its decreased phosphorylation. VHL is often found as a component of an E3 ligase complex in which VHL binds its hydroxylated protein targets for ubiquitination and degradation. We examined the potential effect of VHL on TBK1 ubiquitination and did not find TBK1 ubiquitination induced by VHL (Supplementary Fig. S4A and S4B). PPM1B was reported to be a phosphatase for TBK1 (28). In addition, VHL was also shown to bind with PPM1B (29). We speculated that VHL might affect PPM1B binding with TBK1, therefore contributing to TBK1 dephosphorylation. To test this hypothesis, firstly we overexpressed VHL and found that VHL overexpression led to increased binding between TBK1 and PPM1B (Fig. 2J), which corresponded with decreased TBK1 phosphorylation. Next, we depleted VHL by two independent sgRNAs and found that VHL depletion led to decreased binding between TBK1 and PPM1B, which contributed to increased TBK1 phosphorylation (Fig. 2K). Accumulatively, our data suggest that EglN1 can promote TBK1 hydroxylation, VHL binding and recruitment of PPM1B, which contributes to the dephosphorylation of TBK1 and its decreased activity.
Loss of TBK1 Selectively Suppresses VHL Null ccRCC Cell Growth
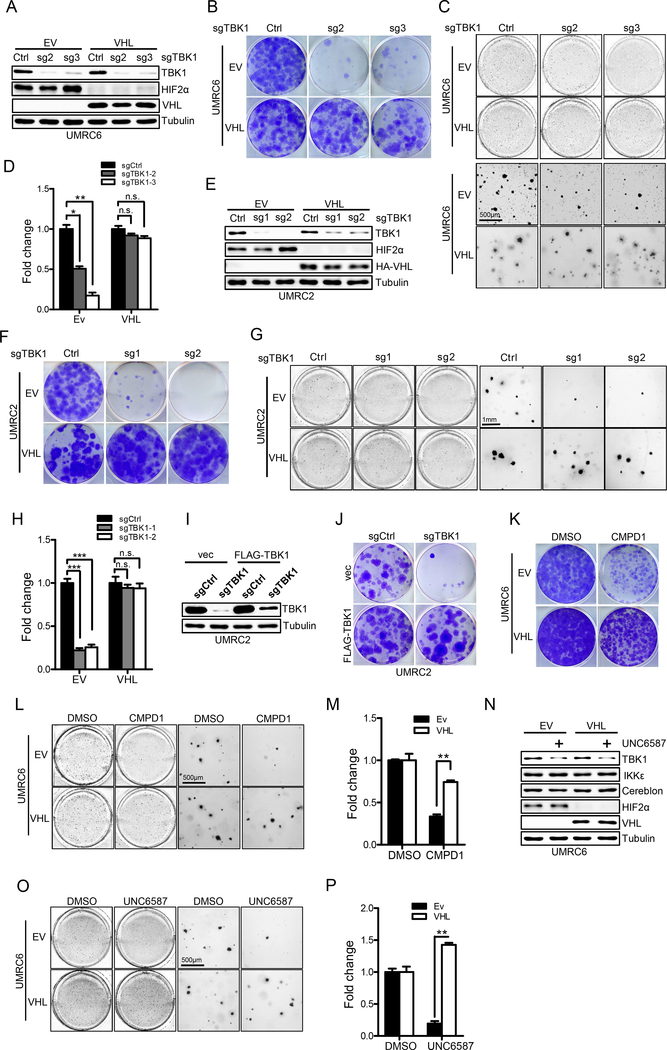
Since we showed that VHL loss led to TBK1 hyperactivation, we next aimed to see whether TBK1 depletion would cause synthetic lethality towards VHL null ccRCC cells. To this end, we depleted TBK1 expression by using several independent sgRNAs in several ccRCC cell lines that are either VHL null (empty vector, EV) or with VHL restoration. In UMRC6 cells, CRISPR-Cas9 mediated TBK1 depletion by sgRNAs led to efficient TBK1 depletion in the isogenic cell lines (Fig. 3A). It is important to note that VHL restoration in VHL null ccRCC cells did not affect cell proliferation and colony formation in serum-rich medium, which is consist with previously published literature (30). Remarkably, TBK1 depletion led to profound cell proliferation defects in VHL null cells whereas VHL restored cells remained largely unaffected (Fig. 3B). We also confirmed this phenotype with 3-D soft agar growth (Fig. 3C–D) and performed these experiments in UMRC2 and RCC4 cells and got similar results (Fig. 3E–H; Supplementary Fig. S5A and S5B). We then depleted TBK1 expression in VHL intact cell lines such as 293T and HKC and found no robust growth defect could be observed (Supplementary Fig. S5C and S5D). To ensure the effect of TBK1 sgRNA was due to its on-target effect on TBK1, we also restored TBK1 expression with the TBK1 construct that carries the silent mutation on sgRNA recognition site and found that TBK1 restoration rescued cell proliferation defect induced by TBK1 depletion (Fig. 3I and J), demonstrating the on-target effect of the TBK1 sgRNA.
Figure 3. Loss of TBK1 selectively suppresses VHL null ccRCC cell growth.
A-D, Immunoblots of lysates (A), crystal violet staining (B), 3-D soft agar growth pictures (C) and quantification of colony numbers (D, duplicate wells) from UMRC6EV/VHL cells infected with lentivirus encoding either Ctrl sgRNA or TBK1 sgRNA.
E-H, Immunoblots of lysates (E), crystal violet staining (F), representative 3-D soft agar growth (G) and quantification of cell colony numbers (H, triplicate wells) of UMRC2EV/VHL cells infected with lentivirus encoding either Ctrl sgRNA or TBK1 sgRNA.
I-J, Immunoblots of lysates and crystal violet staining of UMRC2 cells infected with lentivirus encoding sgCtrl, sgTBK1 (sg2) or pLenti6-vec, pLenti6-FLAG-TBK1 as indicated.
K-M, Representative crystal violet staining (K), 3-D soft agar growth pictures (L) and quantification of colony numbers (M, duplicate wells) of UMRC6EV/VHL cells treated with 4μM CMPD1.
N-P, Immunoblots of lysates (N), representative 3-D soft agar growth pictures (O) and quantification of colony numbers (P, duplicate wells) of UMRC6EV/VHL cells treated with DMSO or 3μM TBK1 PROTAC (UNC6587) as indicated.
Error bars represent SEM, *P<0.05, **P<0.01, ***P<0.001, n.s. denotes no significance.
To further determine whether the effect of TBK1 loss on these phenotypes was dependent on its enzymatic activity, we expressed TBK1 catalytically dead mutant K38A (31) and found that this mutant displayed cell proliferation defects as compared to WT TBK1 (Supplementary Fig. S5E), suggesting that TBK1 enzymatic activity is important for cell proliferation. We also treated UMRC6 and UMRC2 EV or VHL expressing cells with Compound 1 (CMPD1), a recently developed TBK1 inhibitor (32) and found that CMPD1 efficiently blocked soft agar colony growth of EV cells but not VHL restored cells (Fig. 3K–M; Supplementary Fig. S5F and S5G). In addition to CMPD1, we also utilized two previously published TBK1 inhibitors, BX795 and MRT67307 (33,34). We also found that these TBK1 inhibitors preferentially inhibited ccRCC cell growth when VHL is lost and they only modestly affected these cells upon VHL restoration (Supplementary Fig. S5H). UMRC2 cells are resistant to PT2399, a recent reported HIF2α inhibitor (10). We wondered whether there is any cooperative effect between TBK1 inhibitor and PT2399. To answer this question, we treated UMRC2 cells with CMPD1 and PT2399. Compared to single treatment, we didn’t find any cooperative effect between these two compounds (Supplementary Fig. S5I and S5J), which is consistent with the recent publication showing that CDK4/6 inhibitor did not enhance the HIF2α inhibitor efficacy in HIF2α inhibitor resistant cell lines (35).
To eliminate the potential off-target effect associated with these inhibitors, we also generated a novel Cereblon-based TBK1 proteolysis targeting chimera (PROTAC) based on the previous literature (named UNC6587, Supplementary Fig. S6A) since previously reported TBK1 PROTACs facilitated TBK1 degradation via the recruitment of VHL E3 ligase (36,37). Treatment with UNC6587 efficiently eliminated TBK1 protein levels in ccRCC cells, while not affecting its close family member IKKε (Supplementary Fig. S6B and S6C). More importantly, although TBK1 PROTAC efficiently degraded TBK1 in both VHL null and VHL WT ccRCC cells in a similar fashion (Fig. 3N; Supplementary Fig. S6D), TBK1 degradation only caused cell growth defect on 3-D soft agar in VHL null cells (Fig. 3O and P; Supplementary Fig. S6E and S6F), while leaving VHL WT cells unaffected. Therefore, our data suggest that TBK1 may be a therapeutic target in ccRCC with VHL loss.
Loss of TBK1 Inhibits VHL Null ccRCC Tumor Growth
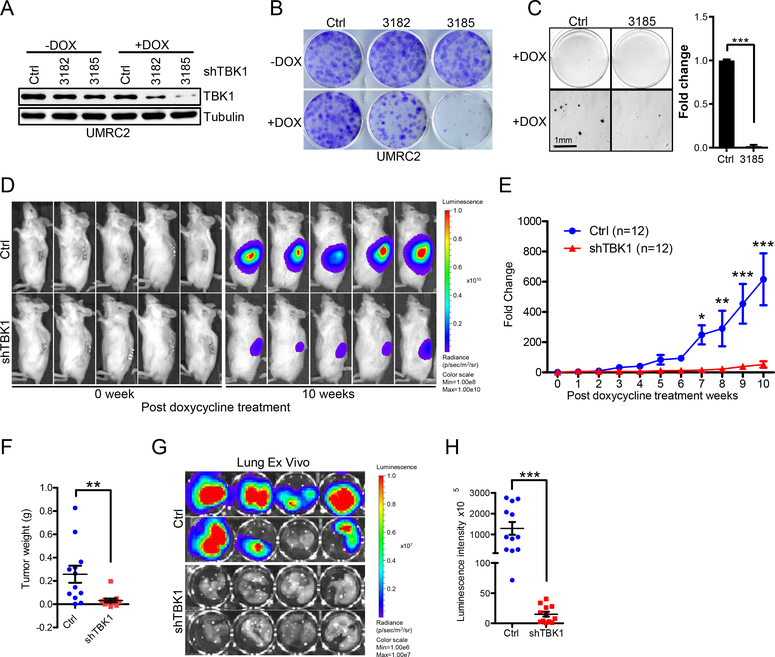
In addition, we examined whether TBK1 was important for maintaining ccRCC tumor growth by introducing two inducible TBK1 shRNAs into UMRC2 cells. These hairpins efficiently depleted TBK1 levels upon doxycycline addition (Fig. 4A). TBK1 depletion led to decreased cell proliferation and reduced soft-agar growth upon doxycycline addition (Fig. 4B and C). Next, either control or TBK1 shRNA (#3185) cells were orthotopically injected into the renal capsules of NSG mice. Upon confirmation of tumor growth in vivo by consecutive weekly bioluminescence imaging, we fed mice doxycycline to induce TBK1 hairpin expression and imaged mice to monitor kidney tumor growth. Whereas cells expressing control hairpin grew readily 10 weeks after addition of doxycycline, TBK1 hairpin-expressing cells failed to proliferate in vivo (Fig. 4D–F). Lung ex vivo imaging data showed that depletion of TBK1 also inhibited tumor cells’ spontaneous lung metastasis (Fig. 4G and H). Our accumulative results suggest that TBK1 is important for ccRCC tumorigenesis both in vitro and in vivo.
Figure 4. Loss of TBK1 selectively suppresses VHL null ccRCC tumor growth.
A-C, Immunoblots of lysates (A) from, representative crystal violet staining (B) and 3-D soft agar growth pictures (C) of UMRC2 cells infected with lentivirus encoding either inducible Ctrl shRNA or inducible TBK1 shRNA and then treated with doxycycline as indicated.
D-E, Representative bioluminescence imaging of before (0 week) and 10 weeks post-doxycycline treatment (D) and quantification of post-doxycycline treatment bioluminescence imaging (E) from UMRC2 luciferase stable cells infected with lentivirus encoding either Teton-control (Ctrl) Teton-shTBK1 3185 (shTBK1) injected orthotopically into the renal capsule of NSG mice. Two-way ANOVA analysis was performed for E.
F, Quantification of kidney tumor weight (n=12).
G-H, Representative lung ex vivo bioluminescence imaging (G) and quantification of ex vivo imaging (H, n=12).
Error bars represent SEM, *P<0.05, **P<0.01, ***P<0.001
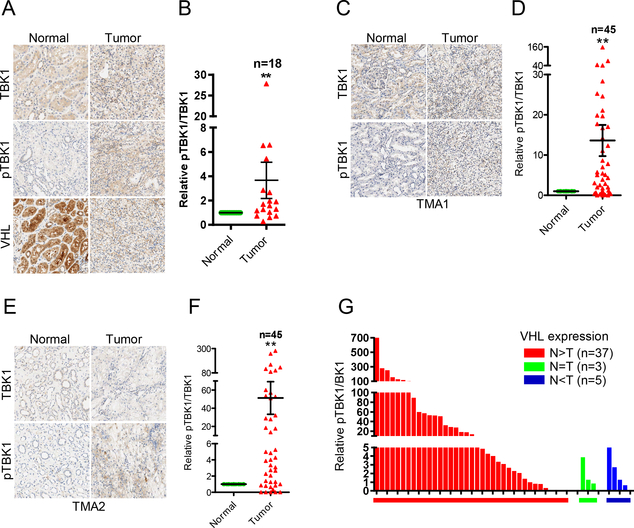
To examine the physiological relevance of TBK1 hyperactivation in ccRCC, we obtained 18 pairs of ccRCC tumors and adjacent normal tissues followed by staining with total TBK1 and phosphorylated TBK1. VHL expression in these tumors was examined by both sequencing and IHC staining. 8 out of 18 tumors were confirmed by sequencing that contain VHL mutations or splice variants (6). The other 10 tumors, although they did not show VHL mutation by sequencing, they still displayed decreased VHL expression by IHC staining (Supplementary Fig. S7), which may be due to epigenetic or other mechanisms. Overall, majority of ccRCC tumors (13 out of 18 pairs) displayed higher TBK1 phosphorylation compared to normal (Fig. 5A and B; Supplementary Table S1). To validate this finding in large clinical cohorts, we also obtained two sets of ccRCC tissue microarrays (TMAs) and stained these TMAs with TBK1 and p-TBK1 with immunohistochemistry (IHC). By calculating the ratio between p-TBK1 and TBK1, there was significant upregulation of p-TBK1 in ccRCC tumors compared to normal in both TMA sets, which was reflected by both representative IHC images and quantitative analyses (Fig. 5C–F; Supplementary Table S1). Annotation information suggested that all tumor samples, except one from TMA2, are ccRCC, which may contain up to 85% of VHL mutations (2). To further confirm VHL loss status, we stained TMA2 with VHL antibody and found 37 out of 45 pairs show decreased VHL level in tumor than paired normal, 3 pairs show comparable level between normal and tumor, while the other 5 tumors have higher VHL than normal tissue. When all samples were divided based on VHL status, we found that tumors express the highest p-TBK1 levels are all from VHL(N>T) group (Fig. 5G). Based on our data, a considerable number of tumors with VHL loss results in no effect on TBK1 phosphorylation. This may result from the complexity of kidney TMAs, including the possessing method, time of procurement and the age of TMAs that might lead to loss of TBK1 phosphorylation in some tumors with VHL loss.
Figure 5. Phosphorylation of TBK1 increases in ccRCC tumor tissues.
A-F, Representative TBK1 and pTBK1 (Ser172) IHC staining pictures and quantification of relative pTBK1/TBK1 signal intensity of tumor and paired normal tissues from a 21-pair ccRCC cohort (A, B), ccRCC TMA1 (C, D) and TMA2 (E, F). Error bars represent SEM, **P<0.01, ***P<0.001 (Wilcoxon matched pairs test).
G, Samples from TMA2 are divided into 3 groups based on VHL expression. Bars represent relative pTBK1/TBK1 ratio of each tumor sample.
TBK1 Phosphorylates p62 on a Bona Fide Ser366 Site and Contribute to ccRCC
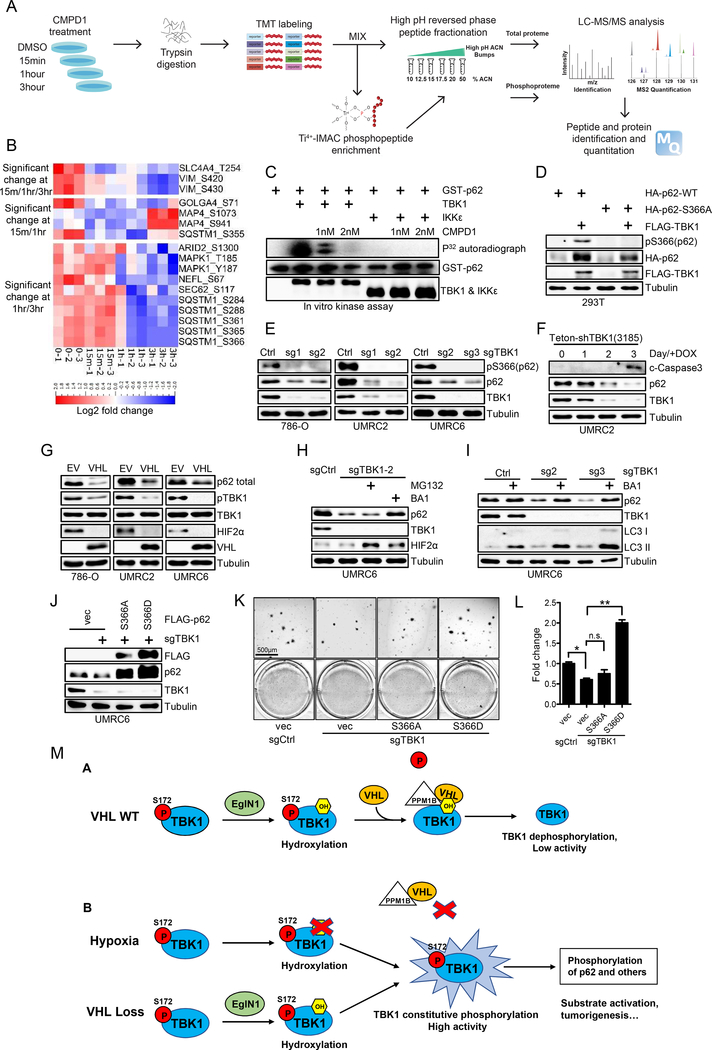
Next, in order to relate kidney cancer associated TBK1 activity with its known innate immune function, we found that we could not detect canonical IRF3 phosphorylation despite detectable IRF3 protein levels (Supplementary Fig. S8A), indicating that there exists a novel mechanism by which TBK1 activation contributes to kidney tumorigenesis. In addition, depletion of STING, an important upstream regulator of TBK1 in innate immunity upon dsDNA infection, did not affect TBK1 protein level or its phosphorylation in multiple ccRCC cell lines (Supplementary Fig. S8B), further strengthening that TBK1 may exert its effect in ccRCC in an innate immunity independent manner. Since Akt is another TBK1 substrate which plays oncogenic function (38,39), we also examined Akt activity by detecting its phosphorylation on Ser473 and Thr308. Our result suggested phosphorylation of Akt Ser473 and Thr308 was not decreased upon TBK1 depletion (Supplementary Fig. S8C). To this end, we performed an unbiased phospho-proteomic screen using UMRC2 cells treated with the specific TBK1 inhibitor CMPD1 for 0, 15min, 1hr and 3 hrs (Fig. 6A). We chose to use the TBK1 inhibitor for the short duration of treatment in the hopes of identifying its direct substrates. We identified 3,320 localized phospho-peptides that are matched to 1243 proteins (Supplementary Fig. S9A). For each treatment, we performed triplicate analysis and we achieved high correlation between replicates. Among differentially expressed phospho-peptides, we are especially interested in the phospho-peptides that were decreased at least at two consecutive time points (either 15 min/1hr, 1hr/3hr or 15min/1hr/3hr). By ANOVA comparison, we obtained a subset of phospho-peptides that showed a significant decrease, defined as p-value<0.05 with log2 fold change≤−0.5 (Fig. 6B). Among this list, we noticed that p62 has been reported to be an oncogene on chromosome 5q that is significantly amplified in kidney cancer (40). By comparing our list with the previous research on the potential p62 phosphorylation sites retrieved from mass spectrometry (41), we found that p62/SQSTM1 Ser366 was the only common site retrieved from both studies. It is interesting to point out that another phosphorylation site on p62 (Ser403) indicated previously in innate immune signaling was not recovered in our list (42), suggesting the context dependent p62 phosphorylation in innate immunity versus certain cancer settings. Therefore, we decided to pursue the potential role of p62 as a novel TBK1 substrate and its phosphorylation (Ser366) in kidney cancer.
Figure 6. TBK1 phosphorylates p62 on Ser366 and stabilize p62 in kidney cancer cells.
A, Schematic of global quantitative phosphoproteomic screen to identify TBK1 substrates in ccRCC cells.
B, Heat map for MS identified phosphorylation sites whose phosphorylation level decreased at 15min/1hr, 1hr/3hr or 15min/1hr/3hr post CMPD1 treatment.
C, Immunoblots and autoradiograph of in vitro kinase assay with hot ATP as indicated.
D, Immunoblots of lysates from 7293T cells transfected with indicated plasmids.
E, Immunoblots of lysates from 786-O, UMRC2 and UMRC6 cells infected with lentivirus as indicated. Cells were collected 3–5 days after puromycin selection.
F, Immunoblots of lysates from UMRC2 cells infected with lentivirus encoding shTBK1(3185) and treated with doxycycline for indicated time.
G-I, Immunoblots of lysates from 786-O, UMRC2 and UMRC6 cells infected with lentivirus as indicated and then treated with MG132 (10μM) or BA1 (10nM) overnight as indicated.
J-L, Immunoblots of lysates (J), representative 3-D soft agar growth pictures (K) and quantification of colony numbers (L, duplicate wells) from UMRC6 cells infected with lentivirus encoding either Ctrl sgRNA or TBK1 sgRNA and then infected with lentivirus encoding FLAG-p62 mutants as indicated. Error bars represent SEM, *P<0.05, **P<0.01, n.s. denotes no significance.
M, Schematic model of hydroxylation dependent regulation of TBK1 activity in normal condition (VHL WT) and VHL loss/hypoxia condition.
Next, we performed an in vitro kinase assay and found that recombinant TBK1, but not IKKε, phosphorylates p62 (Fig. 6C). In addition, CMPD1 inhibits TBK1-induced p62 phosphorylation in a dose dependent manner (Fig. 6C). We then performed the non-radioactive in vitro kinase assay with p62 followed by mass spectrometry and retrieved that p62 Ser366 as the major site being phosphorylated (Supplementary Fig. S9B and S9C). Furthermore, we also confirmed that TBK1 can promote the phosphorylation of p62 on Ser366 site following transfection (Fig. 6D), which can be inhibited by CMPD1 treatment in kidney cancer cells including 786-O and UMRC2 (Supplementary Fig. S9D). Interestingly, by transfecting cells with TBK1, we found that whereas WT p62 level was significantly elevated, p62 S366A mutant displayed diminished upregulation (Fig. 6D). p62 upregulation induced by TBK1 is dependent on TBK1 enzymatic activity since the TBK1 catalytic dead mutant K38A failed to induce p62 upregulation (Supplementary Fig. S9E). Conversely, TBK1 depletion in several ccRCC cell lines (786-O, UMRC2 and UMRC6) all led to decreased p62 protein levels (Fig. 6E). To exclude p62 protein level change is a secondary effect from TBK1 loss induced cell death, we treated inducible TBK1 shRNA with doxycycline in a time course manner (1 to 3 days). We found that, cleaved-caspase3, the marker for cell apoptosis, did not appear apparently until the third day of doxycycline induction, which happened after the decreased TBK1 and p62 levels, suggesting that the regulation of TBK1 on p62 is direct rather than indirect resulting from cell death (Fig. 6F). Our results suggest that TBK1 may directly phosphorylate p62 on Ser366 site and this is important for the stabilization of p62 protein. In accordance with this hypothesis, VHL-null ccRCC restored with exogenous VHL displayed diminished TBK1 phosphorylation and decreased p62 protein levels (Fig. 6G). To determine whether TBK1-depletion induced p62 downregulation was due to proteasomal degradation or lysosomal degradation, we treated these cells with either proteasomal inhibitor MG132 or lysosomal inhibitors including BA1 and NH4Cl and found that the lysosomal but not proteasomal inhibitors significantly elevated p62 expression in TBK1 depleted cells or VHL overexpression cells (Fig. 6H and I; Supplementary Fig. S9F and S9G). Next, to examine whether p62 Ser366 phosphorylation is important for mediating TBK1 knockdown induced phenotype, we overexpressed p62 S366A or S366D in TBK1 sgRNA infected cells and found that p62 S366D could efficiently rescue TBK1 depletion induced cell proliferation defect but S366A couldn’t (Fig. 6J–L). Therefore, TBK1 induced p62 Ser366 phosphorylation is critical to promote ccRCC tumor cell growth.
We also performed p62 IHC staining on 18-pair tumor cohort as well as 2 TMAs. In both TMAs, p62 protein level increased significantly in tumors than paired normal tissues (Supplementary Fig. S10A and S10B; Supplementary Table S1). We then performed correlation analysis but failed to see significant correlation between p-TBK1/TBK1 ratio and p62 protein level. This may be explained by multiple mechanisms that contribute to p62 protein stability (43,44). Especially, it was reported that p62 degradation could be promoted by hypoxia-induced autophagy (45), which may widely exist in cancer cells. Kidney cancer is characterized with chromosome 5q gain as previously published (46), which leads to increased levels of SQSTM1/p62 (40). Therefore, it is likely that tumors with 5q gain may not require increased phosphorylation TBK1 since p62 is hyper-activated in this setting. This may partially explain no correlation between p-TBK1 level and p62 protein level as well. However, it does not rule out the possibility that TBK1 phosphorylation may also contribute to other pathway activation beside p62 that will coordinate with 5q gain to increase kidney tumorigenesis, which will be investigated in future research.
Taken together, our results provide novel mechanistic insight by which TBK1 hyperactivation promotes p62 phosphorylation on Ser366, which lead to upregulation of total and phosphorylation of p62 therefore contributing to ccRCC tumorigenesis.
DISCUSSION
In this study we have uncovered a novel mechanism by which VHL loss or hypoxia promotes TBK1 phosphorylation and activity, therefore promoting its downstream p62 phosphorylation, protein stability and ccRCC tumorigenesis. Under VHL restoration or normoxic condition, TBK1 undergoes EglN1-mediated hydroxylation on Proline 48 residue, which will induce the VHL binding and its recruitment of PPM1B phosphatase to be associated with VHL and TBK1. As a result, PPM1B will de-phosphorylate TBK1 that leads to decreased TBK1 activity. Under hypoxic condition, EglN1 enzymatic activity is inhibited and cannot hydroxylate TBK1 on Proline 48. As a result, similar to VHL loss, TBK1 cannot associate with VHL and PPM1B, which will lead to constitutive TBK1 phosphorylation. As a result of TBK1 hyperactivation, TBK1 will phosphorylate p62 on Ser366 residue and promote renal tumorigenesis (Fig. 6M). Our study characterizes the autonomous function of TBK1 in kidney cancer cells, which is distinctive from its role in innate immune signaling.
VHL loss is a hallmark of clear cell renal cell carcinoma, which accounts for majority of kidney cancer. HIF2α is a well-known oncogene in this setting that has led to clinical trials with the new HIF2α antagonist PT2399. However, only a subset of ccRCC cancer cell lines will display growth inhibition with this HIF2α inhibitor (10,11). In addition, only a subset of ccRCC patients will respond to HIF2α inhibitors and some of them develop resistance to this inhibitor, which could be due to lack of targeting strategies for recently discovered VHL novel substrates including ZHX2 (6). Our study not only adds the TBK1 kinase as novel synthetic lethality partner for VHL null ccRCC, but also puts the TBK1 as a potential therapeutic target for tumor cells that are hypoxic. However, although hypoxia is a characteristic of most solid cancers, it is not necessarily a feature universal to all cancer cells since tumor-wide pseudo-hypoxia is observed in a limited number of cancers, including VHL-deficient ccRCC. The role of TBK1 phosphorylation in other types of cancer cells under hypoxia awaits further detailed investigation.
p62 is an important mediator for tumor cell autophagy since it will facilitate the degradation of specific proteins by autophagy (47). Previous research also showed that VHL null ccRCC displayed high basal levels of autophagy (48). It is interesting that the inhibitors that leads to increased autophagy caused lethality in VHL null ccRCC cells (12). One important consideration is that p62 post-translational modification can modulate the phenotype in VHL null ccRCC cells. Our study firstly demonstrates that p62 Ser366 phosphorylation, mediated by TBK1 kinase, plays an important role on maintaining ccRCC oncogenic phenotype. Our ongoing investigation focuses on whether p62 Ser366 phosphorylation will modulate the autophagy signaling in ccRCC.
Over the decade, research and interest in TBK1 have expanded along with the identification and development of small molecules targeting TBK1. Now at least seven distinct small molecules are known to inhibit TBK1 activity including BX795, compound II, CYT387 (momelotinib), MRT67307, GSK2292978A, amlexanox and CMPD1 (49). However, the problem of selectivity restricts application of these compounds. For example, BX-795 has been reported to inhibit other kinases including PDK1 (33,50). MRT67307 may also inhibit ULK1/2 and block autophagy (51). Only momelotinib and amlexanox have been tested in clinical trials but not for treating kidney cancer (49). Recently developed inhibitor CMPD1, which showed the better selectivity for TBK1 over IKKε (32), was used in our study and showed the selective killing of VHL null ccRCC cells. We have shown in this study that a Cereblon based TBK1 PROTAC may be efficacious in inhibiting VHL-null ccRCC soft agar colony growth but will not affect VHL WT cells. Our ongoing research is to optimize TBK1 PROTACs to achieve higher efficacy and test their effects in tumor xenografts. Recent reports have shown that depletion of TBK1 can boost the efficacy of immune checkpoint inhibitors in cancers (32,52). Therefore, it is likely that by targeting TBK1 kinase, not only can we inhibit the tumor cell autonomous signaling, but also can boost the immunotherapy efficacy in kidney cancer.
METHODS
Cell Culture
RCC4, 786-O, UMRC2, UMRC6, Caki-1, HKC and 293T cells were cultured in DMEM containing 10% fetal bovine serum (FBS) plus 1% penicillin streptomycin. 786-O, Caki-1 and 293T were obtained from ATCC. HKC was obtained from Dr. W. Kimryn Rathmell (53). RCC4, UMCR2 and UMRC6 were obtained from Sigma as described previously(6). Cells were used for experiments within 10–20 passages from thawing. All cells were authenticated via short tandem repeat testing. All cells were tested for ensuring mycoplasma free.
Antibodies and Reagents
Rabbit anti TBK1 (3504), rabbit anti TBK1 phospho-Ser172 (pTBK1, 5483), rabbit anti IKKε (2905), rabbit anti IKKε phospho-Ser172 (pIKKε, 8766), rabbit anti STING (13647), rabbit anti STING phosphor-Ser366 (19781), rabbit anti VHL (68547), rabbit anti EglN1 (3293), rabbit anti HIF1α (3716), rabbit anti p62 (39749), rabbit anti GST (2625), rabbit anti HA tag (3724), rabbit anti FLAG tag (14793), rabbit anti cleaved-caspase3 (9664), mouse anti α-Tubulin (3873) were from Cell Signaling Technology. Mouse anti-HIF2α (ab157249), mouse anti TBK1 (ab12116), mouse anti EglN1 (ab103432), rabbit anti PPM1B (ab70804) were from Abcam. Mouse anti Ub (8017) was from Santa Cruz. Rabbit against p62 phosphor-Ser366 (AF7374) was from Affinity BioSciences. Peroxidase conjugated goat anti-mouse secondary antibody (31430) and peroxidase conjugated goat anti-rabbit secondary antibody (31460) were from Thermo Scientific. DMOG (D1070–1g) was from Frontier Scientific, Deferoxamine (DFO) (D9533–1G), BX-795 (204001–10mg), MRT67307 (506306–5mg) and Bafilomycin-A1 (BA1, B1793) were from Millipore-Sigma, FG4592 (15294–25mg) was from Cayman Chemical. CMPD1 was synthesized by WuxiAPP Tech following the procedure described previously(54).
Identification of Hydroxylated Proline Site on TBK1 by Mass Spectrometry
To prepare samples for mass spectrometry analysis, 293T cells were transfected with FLAG-TBK1 followed by DMSO or 1mM DMOG treatment. Cells were lysed in EBC buffer supplemented with protease inhibitor and phosphatase inhibitor (Roche Applied Bioscience). Then cell lysates were incubated with FLAG M2 beads (Sigma) at 4°C overnight to capture FLAG-TBK1 protein. The next day, beads were washed three times by EBC buffer and the bond proteins were resolved by SDS-PAGE. The gel was stained by Commassie Bright Blue and FLAG-TBK1 bands were excised for mass spectrometry analysis to identify potential hydroxylation site.
Peptide Pull Down Assay
TBK1 peptide containing the WT Pro48 (VFNNISFLRPVDVQMREFE) or hydroxylated Pro48 (VFNNISFLR(P-OH)VDVQMREFE) were synthesized by LifeTein. All peptides were labeled with N-terminal biotin group. To perform pull down assay, 10μg WT or hydroxylated peptides were incubated with 10μl NeutrAvidin beads (Thermo Fisher 29200) for 4hr at 4°C in NETN buffer. After incubation, Avidin beads were washed by NETN buffer to remove free peptides and then incubated with VHL protein generated by in vitro translation system (Promega L1170) in NETN buffer. Then input and bond VHL was examined by Western Blot.
In Vitro Hydroxylation
Purified GST-TBK1 protein (SignalChem T02–10G) was mixed with 10μl EglN1 (in vitro translated) and 75μl hydroxylation buffer (0.5M HEPES pH7.4 containing 10mM FeSO4, 1M Ascorbic Acid, 0.1M α-Ketoglutarate and 1500units/ml Catalase C-100) to make a 100μl reaction system. The reaction was carried out at 37°C for 1hr. After reaction, 500μl NETN buffer and 10μl GST beads (GE Healthcare) were added in and incubated at 4°C for 4hr to pull down GST-TBK1. Then GST beads were washed by NETN buffer to remove unbound proteins and incubated with VHL protein (in vitro translated) overnight. The next day, after 3 times of washing, all input and pull down samples were examined by Western Blot
In Vitro Kinase Assay
For kinase assay using p62 as substrate, 6μl recombinant human p62 protein (Enzo Life Sciences) was mixed with 22μl kinase reaction buffer (20mM Tris-HCl pH7.4, 500mM β-glycerol phosphate, 12mM Magnesium Acetate), 1μl 10mM ATP and 1μl GST-TBK1 (SignalChem) to reach a 30μl reaction system. Kinase reaction was carried out at 30°C, 500rpm for 1hr. After reaction, p62 proteins were resolved by SDS-PAGE and stained by Commassie Bright Blue. The bands were cut out for MS analysis to identify phosphorylation site.
For kinase assay using STING as substrate, 5μl recombinant human STING protein (Active Motif, 81182) was mixed with 33μl kinase reaction buffer (the same as above), 2μl 10mM ATP and 10μl UMRC2 cell lysis to reach a 50μl reaction system. Kinase reaction was carried out at 30°C, 500rpm for 1hr. After reaction, total STING and phospho-STING (Ser366) was detected by Western blot.
Global quantitative phosphoproteomics analysis to identify TBK1 substrates
1. Sample Preparation for Proteomics Analyses
For phosphoproteomics, UMRC2 cells were treated with DMSO or CMPD I for 15 min, 1 hr or 3 hr (n=3 biological replicates per time point). Cells were washed three times with ice-cold cold PBS, then lysed in 8M urea (Sigma, U4883), Tris-HCl (pH 7.6) with protease and phosphatase inhibitors (Roche Applied Bioscience). Lysates were reduced with 5 mM DTT, alkylated with 15 mM iodoacetamide, then subjected to digestion with trypsin (Promega) at a 1:50 enzyme:protein ratio. The resulting peptide samples were acidified and desalted using SepPak C18 SPE cartridges (Waters, 100mg sorbent). Eluates were dried via vacuum centrifugation. Peptide concentration was determined using Pierce Quantitative Colorimetric Peptide Assay. A pooled sample was generated by combining an aliquot (85 μg) of each of the 12 samples. 500 μg of each sample (and 1 mg of pooled sample) was reconstituted with 200 mM TEAB, then individually labeled with a TMT 10-plex reagent (Thermo Fisher) for 5 hr at room temperature. Labeling efficiency and sample ratios were evaluated by LC-MS/MS analysis of two test mixes. Samples were quenched with 50% hydroxylamine to a final concentration of 0.4%. Labeled peptide samples were mixed 1:1 to create two TMT 7-plex sets, dried via vacuum centrifugation, then desalted using SepPak C18 SPE cartridges (Waters, 500mg sorbent). 100 μg of each TMT 7-plex set was fractionated into six ‘global proteome’ fractions using high pH reverse phase spin columns (Pierce). The remaining mixed samples (3 mg of each TMT 7-plex) were enriched for phosphopeptides using MagReSyn Ti-MAC beads (ReSyn Biosciences), as previously described(55). The phosphopeptide-enriched samples were fractionated into three ‘phosphoproteome’ fractions using high pH reverse phase spin columns (Pierce). The proteome and phosphoproteome fractions were dried via vacuum centrifugation and stored at −80°C until further analysis.
2. LC/MS/MS Analysis
The proteome and phosphoproteome fractions were analyzed by LC/MS/MS using an Easy nLC 1200 coupled to a QExactive HF mass spectrometer (Thermo Scientific). Samples were injected onto an Easy Spray PepMap C18 column (75 μm id × 25 cm, 2 μm particle size) (Thermo Scientific) and separated over a 90 min method. For the in-vitro kinase reaction samples, the samples were separated over a 45 min method. The gradient for separation consisted of 5–50% mobile phase B at a 250 nl/min flow rate, where mobile phase A was 0.1% formic acid in water and mobile phase B consisted of 0.1% formic acid in 80% ACN. The QExactive HF was operated in data-dependent mode where the 15 most intense precursors were selected for subsequent HCD fragmentation. Resolution for the precursor scan (m/z 350–1600) was set to 60,000 with a target value of 3 × 106 ions and a maximum injection time of 100 ms. MS/MS scans resolution was set to 60,000 with a target value of 1 × 105 ions and a maximum injection time of 100 ms. Fixed first mass was set to 110 m/z and the normalized collision energy was set to 32% for HCD. Dynamic exclusion was set to 30 s, peptide match was set to preferred, and precursors with unknown charge or a charge state of 1 and ≥ 8 were excluded.
3. Data Analysis
For the proteome and phosphoproteome data, raw data files were processed using MaxQuant version 1.6.1.0., set to ‘reporter ion MS2’ with ‘10plex TMT’. The isolation purity was set to >0.7. Peak lists were searched against a reviewed Uniprot human database (downloaded Feb 2018 containing 20,245 sequences), appended with a common contaminants database, using Andromeda within MaxQuant. All fractions were searched with up to two missed trypsin cleavage sites, fixed Cys carbamidomethylation modification, dynamic Met oxidation and N-terminal acetylation modifications. Peptide false discovery rate was set to 1%. Data were further analyzed and visualized in Perseus, Microsoft Excel and R. Only phosphopeptides identified in both TMT sets with a localization probability score>0.7 were reported. To account for protein levels, log2 intensities were median normalized on both the proteome and phosphopeptide level. The phosphopeptide log2 intensities were normalized to the protein by calculating a ratio using the median normalized protein log2 intensities. ANOVA multiple-sample test was performed in Perseus using a p-value cut-off of 0.05.
For the p62 in-intro kinase assay samples, raw data were processed using Proteome Discoverer 2.1 (Thermo Scientific). Peak lists were searched against a reviewed Uniprot human database, appended with a common contaminants database, using Sequest. The following parameters were used to identify tryptic peptides for protein identification: 10 ppm precursor ion mass tolerance; 0.02 Da product ion mass tolerance; up to two missed trypsin cleavage sites. Carbamidomethylation of Cys was set as a fixed modification and oxidation of Met and phosphorylation of Ser, Thr and Tyr were set as variable modifications. The phosphoRS node was used to localize the sites of phosphorylation. Peptide false discovery rates (FDR) were calculated by the Percolator node using a decoy database search and data were filtered using a 1% FDR cutoff.
Orthotopic Tumor Growth
Six-week old NOD SCID Gamma mice (NSG, Jackson lab) were used for xenograft studies. Approximately 5×105 viable UMRC2 kidney cancer cells were resuspended in 20μl fresh DMEM medium and injected orthotopically into the left kidney of each mice as described previously(40). Bioluminescence imaging was performed as described previously(56,57). For inducible TetOn-TBK1 shRNA, after injection and following consecutive weeks bioluminescence imaging to make sure tumor was successfully implanted in kidney, mice were fed with purina rodent chow #5001 with doxycycline (Research Diets, Inc.). After mice were sacrificed, lung ex vivo imaging was performed immediately to examine tumor metastasis. The rough mass of tumors was presented as mean ± SEM and evaluated statistically using t test. All animal experiments were in compliance with National Institutes of Health guidelines and were approved by the University of North Carolina at Chapel Hill Animal Care and Use Committee.
Statistical Analysis
Unless indicated, the unpaired two-tail student’s t-test was used for experiments comparing two sets of data. All graphs depict mean ± SEM unless otherwise indicated. Graphs were generated by GraphPad Prism. *, ** and *** denotes p value of <0.05, 0.01 and 0.001 respectively. n.s. denotes no significance.
Supplementary Material
SIGNIFICANCE.
The mechanisms that lead to TBK1 activation in cancer and whether this activation is connected to its role in innate immunity remain unclear. Here we discover that TBK1, distinctive from its role in innate immunity, is activated by VHL loss or hypoxia in cancer.
Acknowledgements
We thank UNC LCCC Tissue Procurement Facility, UNC Animal Studies Core and UNC Translational Pathology Laboratory for excellent helps. We thank Prof. Dr. Michael Kracht, Dr. Samuel Bakhoum and Dr. Xuewu Zhang for providing FLAG-TBK1, shSTING plasmids and TBK1 structure information. We would like to thank kidney cancer research program (KCRP) at UT Southwestern for providing us materials with testing. This work was supported in part by a Department of Defense Kidney Cancer Research Program Idea Development Award (W81XWH1910813, to Q.Z.), Kidney Cancer Research Alliance (KCCure), Cancer Prevention and Research Institute of Texas (CPRIT, RP190058 to Q.Z) and the National Cancer Institute (Q.Z., R01CA211732 and R21CA223675; A.B., R35CA197684). Q.Z is an American Cancer Society Research Scholar, V Scholar, Kimmel Scholar, Susan G. Komen Career Catalyst awardee and Mary Kay Foundation awardee. All authors declare no conflict of interest. This research is based in part upon work conducted using the UNC Proteomics Core Facility, which is supported in part by P30 CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center.
Financial support: This work was supported in part by a Department of Defense Kidney Cancer Research Program Idea Development Award (W81XWH1910813, to Q.Z.), Kidney Cancer Research Alliance (KCCure), Cancer Prevention and Research Institute of Texas (CPRIT, RP190058) and the National Cancer Institute (Q.Z., R01CA211732 and R21CA223675; A.B., R35CA197684). Q.Z is an American Cancer Society Research Scholar, V Scholar, Kimmel Scholar, Susan G. Komen Career Catalyst awardee and Mary Kay Foundation awardee.
Footnotes
Conflict of interests: The authors declare no conflict of interests.
REFERENCES
- 1.Godwin JL, Zibelman M, Plimack ER, Geynisman DM. Immune checkpoint blockade as a novel immunotherapeutic strategy for renal cell carcinoma: a review of clinical trials. Discovery medicine 2014;18(101):341–50. [PubMed] [Google Scholar]
- 2.Kaelin WG Jr. Molecular basis of the VHL hereditary cancer syndrome. Nat Rev Cancer 2002;2(9):673–82. [DOI] [PubMed] [Google Scholar]
- 3.Haase VH. The VHL tumor suppressor in development and disease: functional studies in mice by conditional gene targeting. Semin Cell Dev Biol 2005;16(4–5):564–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 2001;292(5516):464–8 doi 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 5.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, et al. Targeting of HIFalpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 2001;292(5516):468–72. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J, Wu T, Simon J, Takada M, Saito R, Fan C, et al. VHL substrate transcription factor ZHX2 as an oncogenic driver in clear cell renal cell carcinoma. Science 2018;361(6399):290–5 doi 10.1126/science.aap8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kondo K, Kim WY, Lechpammer M, Kaelin WG Jr. Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol 2003;1(3):E83 doi 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaelin WG Jr. How oxygen makes its presence felt. Genes & development 2002;16(12):1441–5 doi 10.1101/gad.1003602. [DOI] [PubMed] [Google Scholar]
- 9.Yang H, Kaelin WG Jr. Molecular pathogenesis of the von Hippel-Lindau hereditary cancer syndrome: implications for oxygen sensing. Cell Growth Differ 2001;12(9):447–55. [PubMed] [Google Scholar]
- 10.Cho H, Du X, Rizzi JP, Liberzon E, Chakraborty AA, Gao W, et al. On-target efficacy of a HIF-2α antagonist in preclinical kidney cancer models. Nature 2016;539(7627):107–11 doi 10.1038/nature19795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W, Hill H, Christie A, Kim MS, Holloman E, Pavia-Jimenez A, et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature 2016;539(7627):112–7 doi 10.1038/nature19796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turcotte S, Chan DA, Sutphin PD, Hay MP, Denny WA, Giaccia AJ. A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer cell 2008;14(1):90–102 doi 10.1016/j.ccr.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolff NC, Pavia-Jimenez A, Tcheuyap VT, Alexander S, Vishwanath M, Christie A, et al. High-throughput simultaneous screen and counterscreen identifies homoharringtonine as synthetic lethal with von Hippel-Lindau loss in renal cell carcinoma. Oncotarget 2015;6(19):16951–62 doi 10.18632/oncotarget.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakraborty AA, Nakamura E, Qi J, Creech A, Jaffe JD, Paulk J, et al. HIF activation causes synthetic lethality between the VHL tumor suppressor and the EZH1 histone methyltransferase. Science translational medicine 2017;9(398) doi 10.1126/scitranslmed.aal5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan DA, Sutphin PD, Nguyen P, Turcotte S, Lai EW, Banh A, et al. Targeting GLUT1 and the Warburg effect in renal cell carcinoma by chemical synthetic lethality. Science translational medicine 2011;3(94):94ra70 doi 10.1126/scitranslmed.3002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson JM, Nguyen QH, Singh M, Pavesic MW, Nesterenko I, Nelson LJ, et al. Rho-associated kinase 1 inhibition is synthetically lethal with von Hippel-Lindau deficiency in clear cell renal cell carcinoma. Oncogene 2017;36(8):1080–9 doi 10.1038/onc.2016.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bommi-Reddy A, Almeciga I, Sawyer J, Geisen C, Li W, Harlow E, et al. Kinase requirements in human cells: III. Altered kinase requirements in VHL−/− cancer cells detected in a pilot synthetic lethal screen. Proceedings of the National Academy of Sciences of the United States of America 2008;105(43):16484–9 doi 10.1073/pnas.0806574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barber GN. STING: infection, inflammation and cancer. Nat Rev Immunol 2015;15(12):760–70 doi 10.1038/nri3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 2015;347(6227):aaa2630 doi 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 20.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462(7269):108–12 doi 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper JM, Ou YH, McMillan EA, Vaden RM, Zaman A, Bodemann BO, et al. TBK1 Provides Context-Selective Support of the Activated AKT/mTOR Pathway in Lung Cancer. Cancer research 2017;77(18):5077–94 doi 10.1158/0008-5472.CAN-17-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chien Y, Kim S, Bumeister R, Loo YM, Kwon SW, Johnson CL, et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 2006;127(1):157–70 doi 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 23.Cruz VH, Arner EN, Du W, Bremauntz AE, Brekken RA. Axl-mediated activation of TBK1 drives epithelial plasticity in pancreatic cancer. JCI Insight 2019;4(9) doi 10.1172/jci.insight.126117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou T, Erber L, Liu B, Gao Y, Ruan HB, Chen Y. Proteomic analysis reveals diverse proline hydroxylation-mediated oxygen-sensing cellular pathways in cancer cells. Oncotarget 2016;7(48):79154–69 doi 10.18632/oncotarget.12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Science signaling 2012;5(214):ra20 doi 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haase VH. Renal cancer: oxygen meets metabolism. Exp Cell Res 2012;318(9):1057–67 doi 10.1016/j.yexcr.2012.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordan JD, Simon MC. Hypoxia-inducible factors: central regulators of the tumor phenotype. Current opinion in genetics & development 2007;17(1):71–7 doi 10.1016/j.gde.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y, Liang L, Fan Y, Sun S, An L, Shi Z, et al. PPM1B negatively regulates antiviral response via dephosphorylating TBK1. Cell Signal 2012;24(11):2197–204 doi 10.1016/j.cellsig.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai Y, Song M, Hakala K, Weintraub ST, Shiio Y. Proteomic dissection of the von Hippel-Lindau (VHL) interactome. Journal of proteome research 2011;10(11):5175–82 doi 10.1021/pr200642c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L, Zhang L, Zhang X, Yan Q, Minamishima YA, Olumi AF, et al. Hypoxia-inducible factor linked to differential kidney cancer risk seen with type 2A and type 2B VHL mutations. Molecular and cellular biology 2007;27(15):5381–92 doi 10.1128/MCB.00282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin J, Xiao Y, Chang JH, Yu J, Hu H, Starr R, et al. The kinase TBK1 controls IgA class switching by negatively regulating noncanonical NF-kappaB signaling. Nat Immunol 2012;13(11):1101–9 doi 10.1038/ni.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenkins RW, Aref AR, Lizotte PH, Ivanova E, Stinson S, Zhou CW, et al. Ex Vivo Profiling of PD-1 Blockade Using Organotypic Tumor Spheroids. Cancer discovery 2018;8(2):196–215 doi 10.1158/2159-8290.CD-17-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark K, Plater L, Peggie M, Cohen P. Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase epsilon: a distinct upstream kinase mediates Ser-172 phosphorylation and activation. The Journal of biological chemistry 2009;284(21):14136–46 doi 10.1074/jbc.M109.000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark K, Takeuchi O, Akira S, Cohen P. The TRAF-associated protein TANK facilitates cross-talk within the IkappaB kinase family during Toll-like receptor signaling. Proceedings of the National Academy of Sciences of the United States of America 2011;108(41):17093–8 doi 10.1073/pnas.1114194108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholson HE, Tariq Z, Housden BE, Jennings RB, Stransky LA, Perrimon N, et al. HIF-independent synthetic lethality between CDK4/6 inhibition and VHL loss across species. Science signaling 2019;12(601) doi 10.1126/scisignal.aay0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu J, Qian Y, Altieri M, Dong H, Wang J, Raina K, et al. Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4. Chemistry & biology 2015;22(6):755–63 doi 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crew AP, Raina K, Dong H, Qian Y, Wang J, Vigil D, et al. Identification and Characterization of Von Hippel-Lindau-Recruiting Proteolysis Targeting Chimeras (PROTACs) of TANK-Binding Kinase 1. Journal of medicinal chemistry 2018;61(2):583–98 doi 10.1021/acs.jmedchem.7b00635. [DOI] [PubMed] [Google Scholar]
- 38.Ou YH, Torres M, Ram R, Formstecher E, Roland C, Cheng T, et al. TBK1 directly engages Akt/PKB survival signaling to support oncogenic transformation. Molecular cell 2011;41(4):458–70 doi 10.1016/j.molcel.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie X, Zhang D, Zhao B, Lu MK, You M, Condorelli G, et al. IkappaB kinase epsilon and TANK-binding kinase 1 activate AKT by direct phosphorylation. Proceedings of the National Academy of Sciences of the United States of America 2011;108(16):6474–9 doi 10.1073/pnas.1016132108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L, Shen C, Nakamura E, Ando K, Signoretti S, Beroukhim R, et al. SQSTM1 is a pathogenic target of 5q copy number gains in kidney cancer. Cancer cell 2013;24(6):738–50 doi 10.1016/j.ccr.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Molecular cell 2011;44(2):279–89 doi 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 42.Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 2012;37(2):223–34 doi 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Komatsu M, Ichimura Y. Physiological significance of selective degradation of p62 by autophagy. FEBS letters 2010;584(7):1374–8 doi 10.1016/j.febslet.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 44.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009;137(6):1062–75 doi 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pursiheimo JP, Rantanen K, Heikkinen PT, Johansen T, Jaakkola PM. Hypoxia-activated autophagy accelerates degradation of SQSTM1/p62. Oncogene 2009;28(3):334–44 doi 10.1038/onc.2008.392. [DOI] [PubMed] [Google Scholar]
- 46.Beroukhim R, Brunet JP, Di Napoli A, Mertz KD, Seeley A, Pires MM, et al. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer research 2009;69(11):4674–81 doi 10.1158/0008-5472.Can-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 2009;137(6):1001–4 doi 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bray K, Mathew R, Lau A, Kamphorst JJ, Fan J, Chen J, et al. Autophagy suppresses RIP kinase-dependent necrosis enabling survival to mTOR inhibition. PloS one 2012;7(7):e41831 doi 10.1371/journal.pone.0041831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cruz VH, Brekken RA. Assessment of TANK-binding kinase 1 as a therapeutic target in cancer. Journal of cell communication and signaling 2018;12(1):83–90 doi 10.1007/s12079-017-0438-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feldman RI, Wu JM, Polokoff MA, Kochanny MJ, Dinter H, Zhu D, et al. Novel small molecule inhibitors of 3-phosphoinositide-dependent kinase-1. The Journal of biological chemistry 2005;280(20):19867–74 doi 10.1074/jbc.M501367200. [DOI] [PubMed] [Google Scholar]
- 51.Petherick KJ, Conway OJ, Mpamhanga C, Osborne SA, Kamal A, Saxty B, et al. Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. The Journal of biological chemistry 2015;290(48):28726 doi 10.1074/jbc.A114.627778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 2017;547(7664):413–8 doi 10.1038/nature23270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hacker KE, Fahey CC, Shinsky SA, Chiang YJ, DiFiore JV, Jha DK, et al. Structure/Function Analysis of Recurrent Mutations in SETD2 Protein Reveals a Critical and Conserved Role for a SET Domain Residue in Maintaining Protein Stability and Histone H3 Lys-36 Trimethylation. The Journal of biological chemistry 2016;291(40):21283–95 doi 10.1074/jbc.M116.739375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jenkins RW, Aref AR, Lizotte PH, Ivanova E, Stinson S, Zhou CW, et al. Ex Vivo Profiling of PD-1 Blockade Using Organotypic Tumor Spheroids. Cancer discovery 2018;8(2):196–215 doi 10.1158/2159-8290.CD-17-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Esnault S, Hebert AS, Jarjour NN, Coon JJ, Mosher DF. Proteomic and Phosphoproteomic Changes Induced by Prolonged Activation of Human Eosinophils with IL-3. Journal of proteome research 2018;17(6):2102–11 doi 10.1021/acs.jproteome.8b00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang J, Wang C, Chen X, Takada M, Fan C, Zheng X, et al. EglN2 associates with the NRF1-PGC1alpha complex and controls mitochondrial function in breast cancer. The EMBO journal 2015;34(23):2953–70 doi 10.15252/embj.201591437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Q, Gu J, Li L, Liu J, Luo B, Cheung HW, et al. Control of cyclin D1 and breast tumorigenesis by the EglN2 prolyl hydroxylase. Cancer cell 2009;16(5):413–24 doi 10.1016/j.ccr.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.