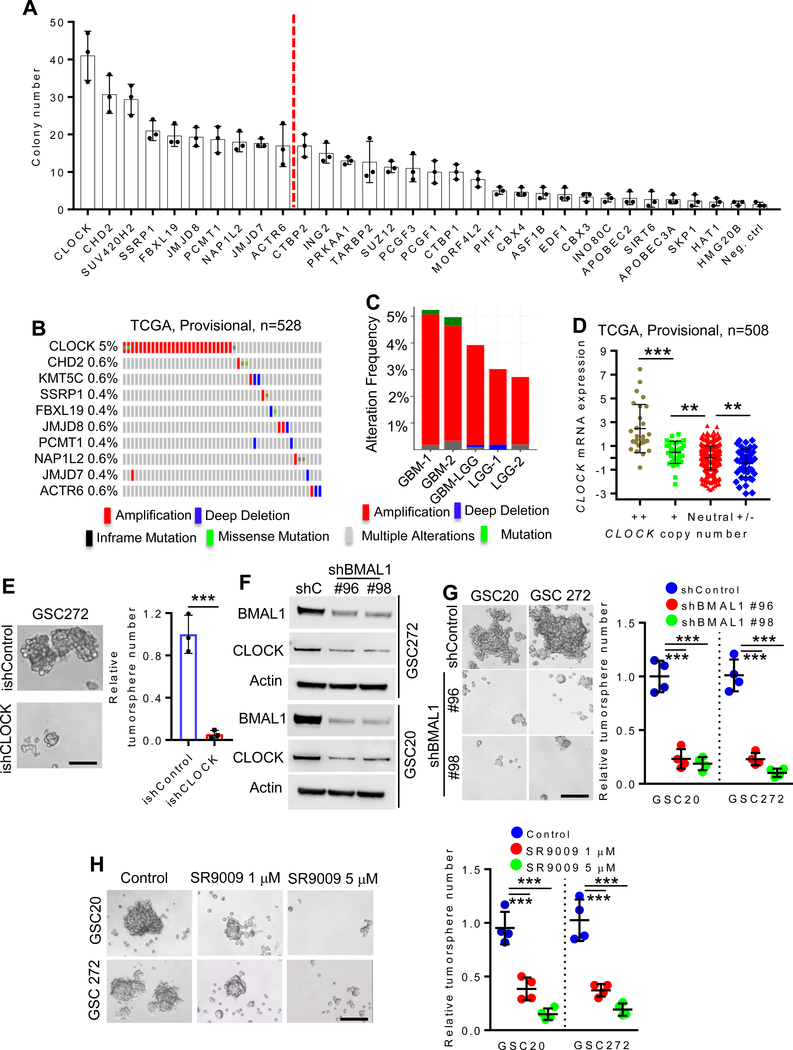
Figure 1. Clock is amplified in GBM and regulates GSC self-renewal.
(A) Soft agar colony formation of hNSCs overexpressing indicated epigenetic genes. n = 3 biological replicates.
(B) Genomic alterations of CLOCK and other epigenetic regulators (CHD2, KMT5C, SSRP1, FBXL19, JMJD8, PCMT1, NAP1L2, JMJD7 and ACTR6) in TCGA GBM database (Provisional dataset; n = 528).
(C) Genomic alteration frequency of CLOCK in TCGA GBM datasets, GBM-LGG merged dataset and LGG datasets.
(D) CLOCK copy number is significantly correlated with CLOCK mRNA expression in TCGA GBM patients (n = 508). ++, high level of amplification; +, gain; Neutral, no change; +/− homozygous deletion. **P <0.01 and ***P <0.001.
(E) Conditional depletion of CLOCK suppresses GBM tumorsphere formation. Representative images (left panel) and quantification (right panel) of tumorspheres in GSC272 cells expressing ishCLOCK or ishControl. n = 3 biological replicates; ***P <0.001.
(F) Immunoblots for CLOCK and BMAL1 in cell lysates of GSC272 and GSC20 expressing shRNA control (shC) or BMAL1 shRNAs.
(G) BMAL1 depletion impairs GSC tumorsphere formation. Representative images (left panel) and quantification (right panel) of tumorspheres in GSC20 and GSC 272 expressing two independent BMAL1 shRNAs or shControl. n = 4 biological replicates; ***P <0.001.
(H) SR9009 treatment impairs GSC tumorsphere formation. Representative images (left panel) and quantification (right panel) of tumorspheres in GSC20 and GSC272 treated with SR9009 at indicated concentrations. n = 4 biological replicates; ***P <0.001.