Abstract
Tumor-associated cell-free DNAs (cfDNAs) are found to play some important roles at different stages of tumor progression; they are involved in the transformation of normal cells and contribute to tumor migration and invasion. DNase I is considered a promising cancer cure, due to its ability to degrade cfDNAs. Previous studies using murine tumor models have proved the high anti-metastatic potential of DNase I. Later circulating cfDNAs, especially tandem repeats associated with short-interspersed nuclear elements (SINEs) and long-interspersed nuclear elements (LINEs), have been found to be the enzyme’s main molecular targets. Here, using Lewis lung carcinoma, melanoma B16, and lymphosarcoma RLS40 murine tumor models, we reveal that tumor progression is accompanied by an increase in the level of SINE and LINEs in the pool of circulating cfDNAs. Treatment with DNase I decreased in the number and area of metastases by factor 3–10, and the size of the primary tumor node by factor 1.5–2, which correlated with 5- to 10-fold decreasing SINEs and LINEs. We demonstrated that SINEs and LINEs from cfDNA of tumor-bearing mice are able to penetrate human cells. The results show that SINEs and LINEs could be important players in metastasis, and this allows them to be considered as attractive new targets for anticancer therapy.
Keywords: circulating cell-free DNA, SINEs, LINEs, tumor, metastasis, horizontal transfer
Introduction
A large amount of data has been accumulated about the composition of circulating cell-free DNA (cfDNA) in the blood of higher organisms, but the biological function of cfDNAs remains to be discussed. Nowadays, several measurements of cfDNA characteristics are used in cancer patient liquid biopsies: SNP analysis of proto-oncogenes and onco-suppressors, changes in the pattern of DNA methylation, and the abundance of oncogene fragments and other specific fragments of tumor-derived DNA microsatellites, tandem repeats, and mobile genetic elements.1, 2, 3 Although attempts have been made to use cfDNAs as indicators of cancer development, based on the content of specific DNA sequences in the blood, the low sensitivity of cfDNA measurement methods, and failure to apply developed markers for early-stage tumor development, has significantly limited the clinical application of this analysis.4,5 On the other hand, increases in microsatellite content, tandem repeats, and mobile genetic elements are commonly detected in the early stages of cancer, both in experimental tumor models and patients, causing researchers to consider these sequences as potential markers for prognosis, as well as factors in disease progression and cure.6,7
Retrotransposons, a type of mobile genetic element (MGE), are able to self-reproduce in the genome through RNA intermediates. These elements can be located in a tandem manner (satellite heterochromatin, telomeres, etc.) or scattered throughout the genome.8 Dispersed short-interspersed nuclear elements (SINEs) are short DNA sequences (less than 500 base pairs) formed by reverse transcription of short RNA molecules: 5S rRNA, tRNA, and various microRNAs (miRNAs).9, 10, 11 SINEs do not encode proteins and their transposition in the genome depends on other mobile elements.12 The most famous and well-investigated SINEs are human Alu-repeats and their murine B-family homologs.13 Long-interspersed nuclear elements (LINEs) are longer retrotransposons (several thousand base pairs in length) containing 3′ end poly(A)-tracts, adenine-rich sequences, or tandem-repeating sequences.14 LINEs are independent elements that encode their own reverse transcriptase (RT) and endonuclease, allowing for genome migration.10
The high abundance of SINEs (particularly Alu) in the blood of cancer patients was first reported in 1977.15 The much higher abundance of LINEs in the cfDNA of cancer patients, compared with healthy donors, was found later, but the described results were controversial.16,17 Nevertheless, researchers have accumulated extensive data on the high levels of SINEs and LINEs, as well as the multiple mutations found in these elements, in the circulating cfDNA of cancer patients and murine models.18, 19, 20, 21 Nowadays, the level of Alu repeats is commonly used to measure the concentration of specific tumor-derived cfDNA in blood serum.22
Despite the extensive data, the role of SINEs and LINEs is still not fully understood, though there is evidence that the elements are regulators that activate onco-genes and suppress onco-suppressors. For example, suppression of the LINE-encoded RT in tumor cells leads to a reduction in tumor progression, both in vitro and in vivo.23 Insertions or deletions of various SINEs and LINEss also lead to the development or suppression of various forms of cancer and other diseases.24, 25, 26, 27 However, the role of the repetitive part of the LINE-element remains unclear, and a reasonable explanation for the role of these elements in carcinogenesis is still unknown. The fact that metastases are often accompanied by a change in the number of SINEs and LINEs in tumor cells, metastatic cells, and the pool of cfDNA requires special attention and additional research.28,29
Previously, DNase I was demonstrated to display high anti-metastatic activity in several tumor models.30, 31, 32, 33 The significant reduction (up to 90%) in the number and area of metastases in Lewis lung carcinoma-bearing mice after treatment with DNase I correlated with a decrease in blood cfDNA concentration and with the restoration of total deoxyribonuclease activity in blood serum to the level of healthy animals.34 Later, we revealed that the molecular targets of DNase I are oncogenes and tandem repeats, including the SINEs and LINEs overrepresented during tumor progression.35
The main goal of our study was to reveal the effect of DNase I on SINEs and LINEs in the pool of circulating cfDNA under the tumor progression in mice and to find the relationship between the level of these tandem repeats and metastasis/tumor development. For these purposes we used three murine models: Lewis lung carcinoma, melanoma B16, and multiple drug resistant (MDR)-positive lymphosarcoma RLS40 (hereafter LLC, B16, and RLS40, respectively). The ability of the SINEs and LINEs derived from the blood serum of tumor-bearing mice to penetrate into tumor cells of another origin was studied to clarify the possibility of horizontal transfer.
Results
Experiment Design
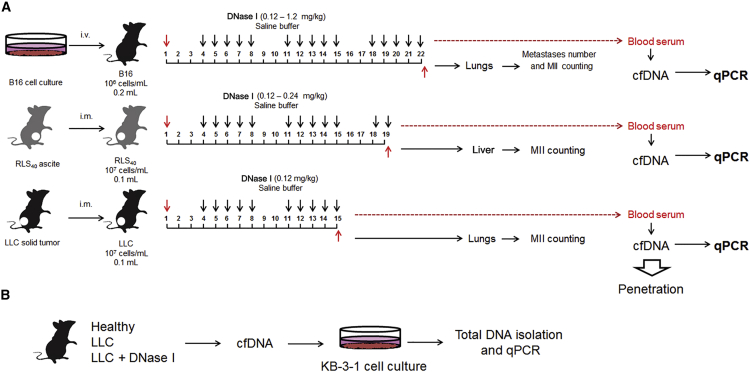
In the first part of our experiments, the anti-metastatic activity of DNase I was studied using three murine tumor models of different histological types, highly relevant to human tumors. Lewis lung carcinoma LLC has epithelial origin and is related to human non-small cell lung cancer.36 Lymphosarcoma RLS40 with a multi-drug resistance phenotype is derived from hematopoietic tissue and is related to the human diffuse large B cell lymphoma.37 Melanoma B16 has neuroectodermal origin and is related to the human metastatic melanoma.38 The experimental scheme is depicted in Figure 1A. The effective dose range of DNase I was chosen on the basis of previously obtained data.34, 35, 36
Figure 1.
Design of the Experiment
(A) The study of the effect of DNase I on inhibition of tumor development and relationship with SINE and LINE level in cfDNA. B16, LLC, or RLS40 cells were transplanted into C57BL/6J mice (B16 and LLC) or CBA mice (RLS40). Starting from the day 4 after the tumor transplantation, animals received intramuscularly saline buffer or DNase I (doses and regiment are indicated on the scheme). i.v., intravenous transplantation, i.m., intramuscular transplantation. (B) The study of the ability of SINEs and LINEs from cfDNA derived from blood of mice with LLC to penetrate KB-3-1 cells of human. Healthy, mice without tumors; w/t, mice with tumors receiving saline buffer; DNase I, mice with tumors receiving DNase I (0.12 mg/kg).
In the process of our study, we evaluated the number of metastases and metastasis inhibition index (MII, see Materials and Methods) in lungs for LLC and B16 models and liver for RLS40 model. cfDNA was isolated from blood serum of tumor-bearing mice 1 h after last injection of DNase I and used for the measurement of the SINE and LINE levels by qPCR.
The second part of our work was devoted to the study of the ability of SINEs and LINEs from the blood of LLC-bearing mice to penetrate human KB-3-1 cells in vitro (Figure 1B). The human epidermoid carcinoma KB-3-1 cell line was chosen to evaluate not only the ability of blood-derived cfDNA to penetrate into the cells, but also the possibility of interspecies horizontal transfer of DNA. KB-3-1 cells were incubated with cfDNA derived from blood of LLC-bearing mice for 3 days. Surface-bound cfDNA were removed to exclude them from the analysis, total DNA was isolated, and the level of SINEs and LINEs was measured. For detailed information, see Materials and Methods.
Evaluation of Anti-tumor and Anti-metastatic Activity of DNase I
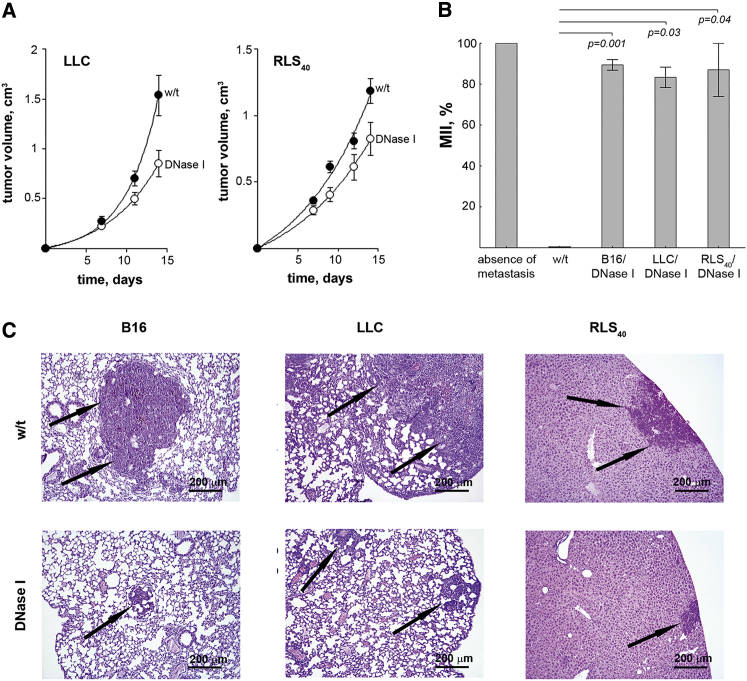
At the beginning of our study, we used the dose of DNase I 0.12 mg/kg that corresponded the dose demonstrated the most antimetastatic effect in previous studies.34,36 Intramuscular (i.m.) treatment of tumor-bearing mice with DNase I affected both primary tumor growth and metastatic spreading (Figure 2). As shown in Figure 2A, mice with LLC and RLS40, treated with DNase I tended to show a decrease in tumor volume: we found a 2-fold decrease in tumor volume for LLC (from 1.6 ± 0.2 in control to 0.8 ± 0.15 cm3 in experimental group) and a 1.5-fold decrease for RLS40 (from 1.2 ± 0.1 in control to 0.8 ± 0.1 cm3 in experimental group) (Figure 2A). The tumor growth inhibition (TGI) was calculated as described in the Materials and Methods section, and was 45% for LLC and 33% for RLS40, respectively.
Figure 2.
Inhibition of Metastases in Mice with LLC, RLS40, and B16 under the Treatment with DNase I
(A) Inhibition of primary tumor growth by DNase I (0.12 mg/kg). Data were statistically analyzed using Student’s t test. Statistical significance p < 0.05. (B) Metastasis inhibition index (MII) in the control and experimental groups. MII = ([mean metastasis area control – mean metastasis area experiment]/ mean metastasis area control) × 100%. Data were statistically analyzed using one-way ANOVA with a post hoc Tukey test. Data are presented as mean ± SE. Statistical significance p < 0.05. (C) Representative histological images of mouse organs with metastases (lungs and liver) treated with DNase I. Metastases are indicated by black arrows. Hematoxylin and eosin staining, original magnification ×400. w/t, mice with tumors receiving saline buffer; DNase I, mice with tumors receiving DNase I (0.12 mg/kg).
In all experimental groups of tumor-bearing mice, the administration of DNase I resulted in a significant decrease in the number of surface metastases in the lungs (B16 model) and a decrease in the area of internal metastases in the lungs (LLC and B16 models) and liver (RLS40 model), in comparison with the control animals. Inhibition of surface metastases development was assessed by counting metastases on the lung surface. In the control group with melanoma B16, the average number of surface lung metastatic foci was 117 ± 21, while in animals treated with DNase I it was 61 ± 11 (primary data not shown).
Inhibition of internal metastases development was assessed by counting the average metastases area and metastasis inhibition index (MII) using morphometric analysis (Figure 2B). The MII of the control group was taken as 0%, and the MII corresponding to 100% indicated the absence of metastases. We observed a 6-fold decrease in the internal lung metastasis area for LLC (p = 0.009), a 7.7-fold decrease in the internal liver metastasis area for RLS40 (p = 0.003), and a 9.4-fold decrease in the internal lung metastasis area for melanoma B16 (p = 0.0002). MII was calculated on the basis of meanings of internal metastases area. Histological and morphometric analysis revealed that the MII of mice with LLC, RLS40, and melanoma B16 treated with DNase I was approximately the same within the groups and amounted to 83%–89% on average (Figure 2B).
Morphometry of cross sections of lungs and liver (Figure 2C) show that DNase I administration evidently reduces the average metastases area. In histological studies, melanoma B16 in pulmonary metastatic foci was represented by atypical polymorphic and spindle-shaped cells containing the brown pigment melanin (Figure 2C, left panel). Primary tumor node and pulmonary metastatic foci of LLC consisted of polymorphic cells with a rounded shape and a large nucleus with large nucleoli and condensed chromatin grains (Figure 2C, middle panel). RLS40 primary tumor node and metastatic foci in the livers were represented by monomorphic atypical lymphoid cells with frequent mitosis (Figure 2C, right panel).
The Effect of DNase I on the Concentration of cfDNA in Blood of Tumor-Bearing Mice
cfDNA was isolated from blood serum and processed, as described in the Materials and Methods. The cfDNA levels in the blood serum of healthy and tumor-bearing mice, before and after the treatment with DNase I, are shown in Table 1. The concentration of cfDNA in the blood of healthy C57Bl and CBA mice was 65 ± 10 ng/mL and 168 ± 32 ng/mL, respectively. Tumor progression in mice treated with saline buffer (control) resulted in a 2-fold increase in cfDNA concentration in the blood of animals with LLC (reached 138 ± 20 ng/mL) and RLS40 (reached 300 ± 91 ng/mL). The most pronounced increase in cfDNA concentration was found for mice with B16; in this case the cfDNA level reached 456 ± 82 ng/mL, which was 5- to 6-fold higher in comparison with healthy animals Table 1). DNase I treatment led to a significant reduction in cfDNA levels in the blood of mice with LLC and B16, but not in the blood of mice with RLS40. We observed a 2-fold and 1.5-fold decrease in cfDNA concentrations after DNase I treatment, for B16 and LLC, respectively.
Table 1.
Primer Sequences Used for Real-Time PCR
| Repeat Type | Primer Sequences 5′-3′ | Amplicon Length |
|---|---|---|
| B1_Mm_F | 5′-TGGCGCACGCCTTTAAT-3′ | 92 |
| B1_Mm_R | 5′-TCCTGGACCTCACTTTGTAGA-3′ | |
| B1_mur4_F | 5′-AGGCGGATTTCTGAGTTCAA-3′ | 91 |
| B1_mur4_R | 5′-GAGACAGGGTTTCTCTGTGTAG-3′ | |
| B1_mus1_F | 5′-AGGCGGATTTCTGAGTTCAA-3′ | 91 |
| B1_mus1_R | 5′-GAGACAGGGTTTCTCTGTGTAG-3′ | |
| B1_mus2_F | 5′-GAGACAGGCGGATTTCTGAGT-3′ | 71 |
| B1_mus2_R | 5′-TGTAGCCCTGGCTGTCCT-3′ | |
| L1_mus 1_F | 5′-GCCAGGTATCTGTGCATCTT-3′ | 87 |
| L1_mus 1_R | 5′-ACTCTAGCTCTCTCCTGAGTTT-3′ | |
| L1Md_F_F | 5′-GCTACTATACCGAGCCAAACTC-3′ | 99 |
| L1Md_F_R | 5′-GCTGGATTCGTGGAGAGATAAT-3′ | |
| L1Md_Gf_F | 5′-GCTACTATACCCAGCCAAACTC-3′ | 91 |
| L1Md_Gf_R | 5′-CGTGGAGAGATAATGCGTGAA-3′ | |
| Lx2_mus1_F | 5′-ACATGGTATGTGCTCACTGATAA-3′ | 96 |
| Lx2_mus1_R | 5′-CTCTTCCTTCTTGGGCTTCAT-3′ |
Analysis of Abundance of SINEs and LINEs in Blood of Tumor-Bearing Mice
Recently, we observed an increase in the representation of many types of tandem repeats upon LLC development (sequencing data35). These tandem repeats are referred to as mobile genetic elements, such as retrotransposons, and most of the repeats are related to LINEs and SINEs.35 Treatment of LLC-bearing mice with DNase I caused a drop in the content of these elements in the bloodstream, thus showing that the anti-metastatic effect of DNase I could be executed by changing the profile of cfDNA, particularly the level of LINEs and SINEs. In other words, DNase I may target these elements.
To evaluate the abundance of SINEs and LINEs in the blood of mice with tumors of various origins and to clarify their role in metastases dissemination, we performed PCR analysis of SINEs and LINEs in cfDNA isolated from the blood of healthy mice, control tumor-bearing mice, and those treated with DNase I.
For the analysis, we chose four SINEs and four LINEs well represented in the sequencing data, whose level significantly increased upon LLC development and dropped after DNase I treatment.35 Four types of repeats belong to B1 subfamily (B1_Mm, B1_mur4, B1_mus1, and B1_mus2), three types of repeats belong to L1 subfamily (L1_mus 1, L1Md_F, and L1Md_Gf), and one type belongs to Lx subfamily (Lx2_mus 1). PCR primers for these repeats were selected for the first time (see Materials and Methods, Table 2).
Table 2.
The cfDNA Levels in the Blood Serum of Healthy Mice and Mice with Tumors (B16, LLC, RLS40) Before and After the Treatment with DNase I
| Mice | Healthy, ng/mL | Tumor Type | w/t, ng/mL | DNase I, ng/mL |
|---|---|---|---|---|
| C57Bl | 65 ± 10 | B16 | 456 ± 82 | 246 ± 22 |
| LLC | 138 ± 20 | 97 ± 16 | ||
| CBA | 168 ± 32 | RLS40 | 300 ± 91 | 292 ± 89 |
w/t, mice with tumors receiving saline buffer; DNase I, mice with tumors receiving DNase I (0.12 mg/kg). Data were statistically analyzed using one-way ANOVA with a post hoc Tukey test. Data are presented as mean ± SE. Statistical significance p < 0.05. Statistical differences between groups p < 0.0001.
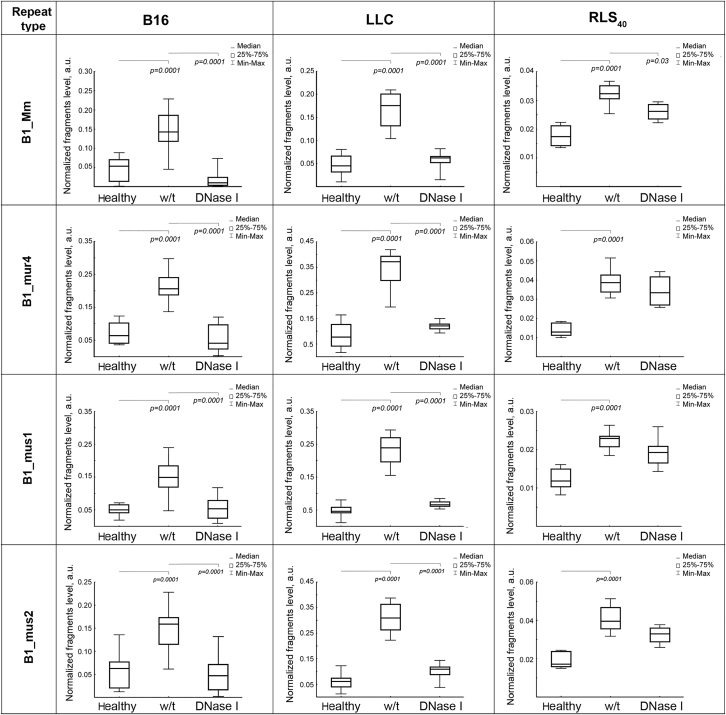
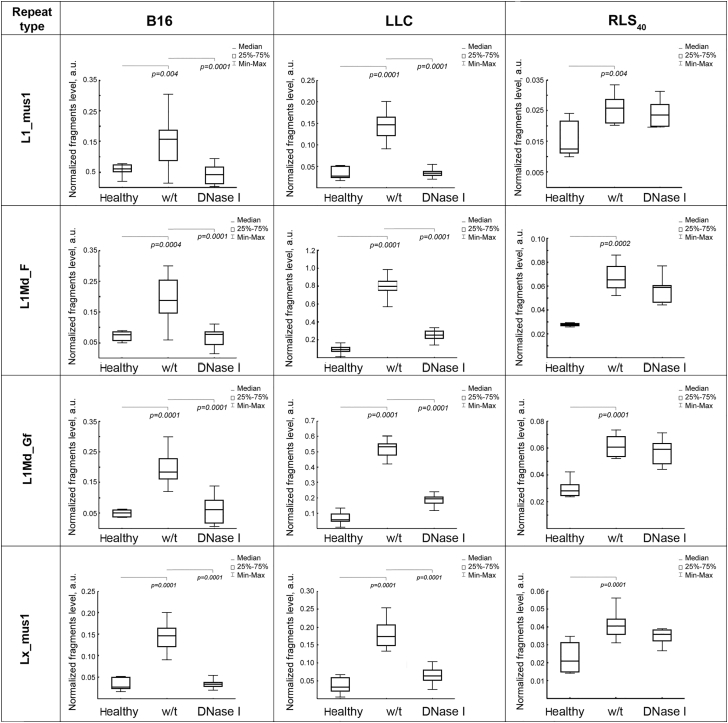
The levels of SINE and LINE fragments in cfDNA samples from different groups of mice were detected using real-time PCR. The PCR meanings were normalized to the concentration of cfDNA and volume of blood serum and expressed as arbitrary unit (a.u.). Normalized levels of SINEs and LINEs in the cfDNA samples from healthy mice, control tumor-bearing mice (w/t), and those treated with DNase I are shown in Figures 3 and 4.
Figure 3.
Boxplots of the Level of SINEs B1_Mm, B1_mur4, B1_mus1, and B1_mus2 among cfDNA in the Blood Serum of Healthy Mice, Mice with B16, LLC, or RLS40 Before and After the Treatment with DNase I
Data of qPCR. The fragment level is presented as arbitrary unit (a.u.). Data were statistically analyzed using one-way ANOVA with a post hoc Tukey test. Boxes represent 25th, 50th, and 75th percentiles. Squares with line represent median. Whiskers represent minimum/maximum. Statistical significance p < 0.05. Healthy, mice without tumors; w/t, mice with tumors receiving saline buffer; DNase I, mice with tumors receiving DNase I (0.12 mg/kg).
Figure 4.
Boxplots of the Level of LINEs L1_mus1, L1Md_F, L1Md_Gf, and Lx_mus1 among cfDNA in the Blood Serum of Healthy Mice, Mice with B16, LLC, or RLS40 Before and After the Treatment with DNase I
Data of qPCR. The fragment level is presented as a.u. Data were statistically analyzed using one-way ANOVA with a post hoc Tukey test. Boxes represent 25th, 50th, and 75th percentiles. Squares with line represent median. Whiskers represent minimum/maximum. Statistical significance p < 0.05. Healthy, mice without tumors; w/t, mice with tumors receiving saline buffer; DNase I, mice with tumors receiving DNase I (0.12 mg/kg).
In the cfDNA of healthy mice, the SINEs and LINEs abundance was very low and varied depending on mouse line. In the blood of healthy C57BL/6 mice, the level of these elements was 0.05–0.08 a.u. (Figures 3 and 4, panels B16 and LLC). cfDNA of CBA mice was characterized by a lower level of all SINEs (0.012–0.017 a.u.) and two LINEs (0.015 for L1_mus1 and Lx_mus1), whereas two other LINEs (L1Md_F and L1Md_Gf) were presented in amounts close to that found in the blood of healthy C57BL/6 mice (0.025; Figure 4, panel RLS40).
Progression of B16 and LLC tumors is accompanied by a significant increase in the abundance of SINEs and LINEs. In the case of B16 tumors, the level of SINEs increased 3–4 times (Figure 3, panel B16) and the level of LINEs increased 3–3.4 times (Figure 4, panel B16). Similarly, LLC development caused a 3.4- to 5-fold increase in the level of SINEs (Figure 3, panel LLC), while the level of LINEs increased even more significantly in comparison with SINEs: the abundance of LINEs increased from 3 times (for L1_mus1 and Lx_mus1) to 8 times (for L1Md_F) and 12 times (for L1Md_Gf; Figure 4, panel LLC). Progression of RLS40 was also accompanied by a 1.8–2.2 increase in the level of SINEs and LINEs, nevertheless, the level of these elements remained rather low and reached a maximum of 0.04 a.u. for SINEs (Figure 3, panel RLS40).
Treatment of B16- or LLC-bearing mice with DNase I lead to the decrease of SINEs and LINEs level to close to the level of healthy animals (Figures 3 and 4). In the case of RLS40-bearing mice, we observed a tendency for SINEs and LINEs abundance to decrease, but these changes were mainly statistically insignificant, decrease was only statistically significant for B1_Mm (Figures 3 and 4, panel RLS40).
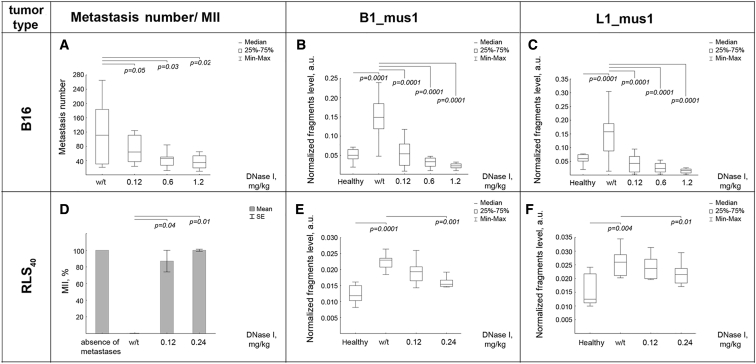
The Effect of DNase I Doses on the Level of SINEs and LINEs in Blood and Metastases in Tumor-Bearing Mice: B16 and RLS40 Models
To investigate the DNase I dose effect, we used B16 and RLS40 tumor models. Tumors were induced and treatment was provided in mice, as described in the Materials and Methods section. The number of lung (B16 tumors) and liver (RLS40 tumors) metastases was determined by counting (B16) and by morphometric analysis (B16 and RLS40, Figure 5). Analysis of surface lung metastases in B16-bearing mice revealed that DNase I caused a decrease in metastases number in a dose-dependent manner. DNase I at a dose of 0.12 mg/kg reduced twice the metastases number (61 ± 11 versus 117 ± 21 in the control, Figure 5A). An increase in the dose of DNase I to 0.6 mg/kg led to a decrease in metastases number to 48 ± 10, however, a further increase of the dose to 1.2 mg/kg did not result in a significant reduction in metastases number (35 ± 5, Figure 5A). It is worth mentioning that, in the RLS40 model, DNase I at the dose of 0.12 mg/kg inhibited primary tumor growth by a factor of 1.5, compared to the control group (Figure 2B), and increasing the dose to 0.24 mg/kg resulted in tumor growth inhibition of a factor of 2.5 (primary data not shown). In the RLS40 model, DNase I at a dose of 0.12 mg/kg led to the inhibition of metastases of up to 87%, and doubling the dose caused an even more significant reduction of metastasis area of up to 100% (Figure 5D).
Figure 5.
Dosage Effect of the DNase I on the Metastasis Number and SINEs/LINEs Levels in Blood Serum of Tumor-Bearing Mice
Mice with implanted B16 or RLS40 tumors received intramuscularly saline buffer (w/t) or DNase I (0.12–1.2 mg/kg) (for details see Figure 1 and section Materials and Methods). (A) The number of surface metastasis for B16 model. (B) The level of B1_mus1 elements for B16 model. (C) The level of L1_mus1 elements for B16 model. (D). MII for RLS40 model. (E) The level of B1_mus1 elements for RLS40 model. (F) The level of L1_mus1 elements for RLS40 model. B1_mus1 and L1_mus1 were analyzed by qPCR. The fragment level is presented as a.u. Data were statistically analyzed using one-way ANOVA with a post hoc Tukey test. (A–C, E, and F) Data are presented as median. Boxes represent 25th, 50th, and 75th percentiles. Squares with line represent median. Whiskers represent minimum/ maximum. (D) Data are presented as mean ± SE. Statistical significance p < 0.05. Healthy, mice without tumors; w/t, mice with tumors receiving saline buffer; DNase I, mice with tumors receiving DNase I in different doses.
As expected, i.m. administration of DNase I caused a dose-dependent decrease in cfDNA concentration in the blood serum of B16-bearing mice. The concentration of cfDNA in the control group (C57Bl mice, melanoma B16) was 456 ± 82 ng/mL, whereas in the mouse groups receiving DNase I in the doses 0.12, 0.6, and 1.2 mg/kg, cfDNA concentrations were 246 ± 22, 92 ± 10, and 83 ± 6 ng/mL, respectively. In contrast to the B16 tumor model, we did not observe dose-dependent decreases of cfDNA concentration in RLS40 tumors: the concentration of cfDNA in the control group was 300 ± 91 ng/mL, whereas both at low doses (0.12–0.24 mg/kg) and high doses (0.6 and 1.2 mg/kg), we found statistically insignificant alterations in concentration averaging 286 ± 93 ng/mL.
The effect of DNase I dose on the content of SINEs and LINEs (B1_mus1 and L1_mus1) was analyzed by qPCR. The B16 progression led to a 3- to 4-fold increase in SINEs and 3- to 5-fold increase in LINEs, from 0.05 ± 0.02 a.u. and 0.06 ± 0.015 a.u. for healthy mice to 0.15 ± 0.045 a.u. and 0.17 ± 0.055 a.u. in B16-bearing mice, respectively (Figures 5B and 5C). DNase I treatment at low doses (0.12 mg/kg) led to a significant reduction of B1_mus1 and L1_mus1 levels in cfDNA, in a dose-dependent manner: the SINE and LINE levels were 0.05 ± 0.03 a.u. and 0.045 ± 0.03 a.u., respectively (Figures 5B and 5C). A further increase in the DNase I dose caused an even lower reduction of both B1_mus1 and L1_mus1 levels, coming close to levels in healthy mice: to 0.03 ± 0.01 a.u. and 0.02 ± 0.01 a.u., respectively (Figures 5B and 5C).
In the case of RLS40, the tumor progression led to a significant increase of SINE and LINE levels, from 0.012 ± 0.003 a.u. and 0.013 ± 0.005 a.u. (healthy mice) to 0.023 ± 0.003 a.u. and 0.026 ± 0.003 a.u., respectively (Figures 5E and 5F). DNase I treatment of RLS40-bearing mice was accompanied by less pronounced, but dose-dependent, effects on SINE and LINE levels in cfDNA, compared with the B16 melanoma model. At a low dose of DNase I, we observed a decrease in B1_mus1 and L1_mus1 levels (0.019 ± 0.002 a.u. and 0.023 ± 0.003 a.u., respectively), but it was statistically insignificant. Only at a dose of 0.24 mg/kg, we detected a significant reduction of B1_mus1 (0.015 ± 0.0015 a.u.) and a slight alteration of L1_mus (0.021 ± 0.0025 a.u.) (Figures 5D and 5F).
Study of the Ability of cfDNA Derived from Blood of Tumor-Bearing Mice to Penetrate KB-3-1 Cells
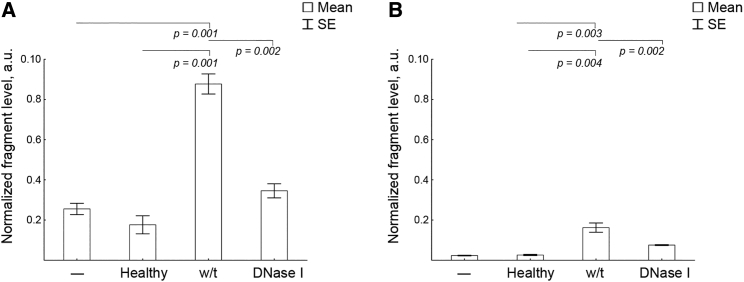
At the beginning, we proved that the baseline level of SINEs and LINEs (B1-mus1 and L1_mus1) in KB-3-1 cells was 105 times lower in comparison to B16, LLC, and RLS40 cells (primary data not shown). The experimental setup is depicted in Figure 1B. KB-3-1 cells were treated with cfDNA and then surface-cell bound DNA was removed to exclude them from the analysis. The total DNA was isolated and B1_mus1 and L1_mus1 levels were detected using qPCR. Normalized levels of SINEs and LINEs are shown in Figures 6B and 6C). No toxic effects of cfDNA on cell proliferation were detected.
Figure 6.
Detection of SINE and LINEs in KB-3-1 Cells after Incubation with cfDNA Derived from Blood of Healthy Mice, Mice with LLC Treated with Saline Buffer, or DNase I
(A) The levels of B1_mus1 SINE. (B) The levels of L1_mus1 LINE. Data of qPCR. The fragment level is presented as a.u. Data were statistically analyzed using one-way ANOVA with a post hoc Tukey test. Data are presented as mean ± SE. Statistical significance p < 0.05. –, intact cells; healthy, cells incubated with blood-derived cfDNA of healthy mice; w/t, cells incubated with blood-derived cfDNA of LLC-bearing mice treated with saline buffer; DNase I, cells incubated with blood-derived cfDNA of LLC-bearing mice treated with DNase I.
We found that treatment of KB-3-1 cells with cfDNA, derived from the blood serum of healthy mice, did not lead to an increase in SINEs and LINEs in total DNA of KB-3-1 cells: the levels of these elements was close to the levels in untreated KB-3-1 cells (Figures 6B and 6C). Treatment of KB-3-1 cells with blood-derived cfDNA from LLC-bearing mice resulted in a 5-fold increase in B1-mus1 elements (Figure 6B). We found an 8-fold elevation in L1-mus1 levels in total DNA of KB-3-1 cells after cell treatment with blood-derived cfDNA from LLC-bearing mice (Figure 6C). Incubating KB-3-1 cells with blood-derived cfDNA of LLC-bearing mice, treated with DNase I, did not increase the level of SINEs in total DNA of KB-3-1 cells; the levels were comparable with those for untransfected cells and cells treated with healthy-derived cfDNA (Figure 6B). As for LINEs, their level was slightly higher than for untransfected cells and cells treated with blood-derived cfDNA of healthy mice; nevertheless, it was low and did not reach 0.01 (Figure 6C). Obtained results correlate with the decrease in SINEs and LINEs in the blood serum of mice with LLC after treatment with DNase I.
Discussion
Previously we demonstrated that the anti-metastatic effect of DNase I is accompanied by an increase in deoxyribonucleic activity in the blood and a reduction in the concentration of circulating cfDNA, and the majority of changes are observed for tandem repeats, including those belonging to B, L1, and Lx families.35 In this work we analyzed the relation between the level of SINEs and LINEs in the blood-derived cfDNA of mice with different metastatic tumors—LLC, melanoma B16, and resistant lymphosarcoma RLS40 having relevance to human tumors37, 38, 39—and the effect of DNase I on primary tumor and metastases.
Today, plasma is more often used both when working with animals and patients, nevertheless we chose blood serum as a source of cfDNA in connection with our previous studies where we analyzed cfDNA profile by NGS. In spite of serum is unavoidably contaminated by genomic DNA in our case this is only but a detail because the use of serum, in combination with the selected cfDNA isolation technique, allows isolating almost completely cfDNA material, including ultrashort sequences. Similarly, in PCR analysis, since we use internal normalization between samples, the contribution of genomic DNA can be neglected.
Our results demonstrated that DNase I affects both primary tumor growth and metastases spreading. In RLS40 and LLC models, DNase I inhibited primary tumor growth by factor 1.5–2 and metastases development by factor 6–9. It should be noted that this is the first time DNase I has been observed to induce such a significant retardation of primary tumor growth in RLS40 and LLC models, although no such effect for LLC model was previously detected that can be explained by different conditions of the experiment.34 It is worth noting that other research did not observe the effect of DNase I on primary tumor node in the xenograft model of human pancreatic32 and lung cancer40 on murine model and glioma on rat model.41 Only when using a mixture of DNase I and proteases (papain, trypsin, and chymotrypsin) authors demonstrate a significant decrease in the size of the tumor node.40 Nevertheless, DNase I was shown to decrease the proliferation rate of tumor cells in vitro33,42 and reduced the concentration of extracellular cfDNA in culture medium.42 Thus, the data on the inhibition of the primary tumor growth by DNase I in vivo are consistent with the data on the effect of DNase I on tumor cell proliferation obtained in vitro.
We found that the development of all three types of tumors is accompanied by both an increase in the total concentration of cfDNA in blood of tumor-bearing mice (Table 1) and an increase in the level of SINEs and LINEs in the blood-derived cfDNA content (Figures 3 and 4). Our data demonstrated that DNase I caused the decrease in the level of SINEs and LINEs in blood of tumor-bearing mice that correlated with the inhibition extent of primary tumor growth and metastases development. We suggest that DNase cleaves cfDNA in the blood in a nonrandom manner, and the observed decrease in the level of SINEs and LINEs may be due to their significantly higher accessibility to cleavage. In B16 and RLS40 models, we observed a dose-dependent effect of DNase I on the number of metastases, which correlated with a decrease in both the total concentration of cfDNA and the abundance of SINEs and LINEs.
The essential increase of SINEs in the blood of mice under tumor development is in agreement with recent data on the elevation of Alu repeats in the blood of patients with non-small cell lung cancer (NSCLC), breast cancer, and ovarian cancer.1,6, 7, 8,43 Despite the fact that overexpression of RT encoded by LINEs has been previously detected in tumors,23,44,45 this is the first demonstration of the alteration of LINE levels in circulating blood-derived cfDNA under tumor progression.
The possible role of SINEs and LINEs in carcinogenesis is currently widely discussed. The data accumulated so far show there is a higher abundance of Alu-fragments in blood-derived cfDNAs of cancer patients, in comparison with healthy donors.46 It has been repeatedly observed that an increase in the number of Alu elements among cfDNA, in the development of many cancers, correlates with disease development stage and severity.21,47 Disruption of gene integrity due to the insertion of Alu-repeats leads to a change in methylation patterns, and transcription of the gene results in the development of diseases, including cancer.10,27 Insertions and deletions of LINEs can also alter the gene expression and lead to unequal recombination and instability of the genome, changes in cell phenotype and the acquisition of malignant cell traits.17,48, 49, 50 There are also data showing that somatic mutations of proto-oncogenes and onco-suppressors, caused by the insertion of retrotransposons, lead to the appearance of tumors.24, 25, 26
The increasing levels of SINEs and LINEs in tumor and their involvement in metastatic spreading is clear. The role of cfDNAs in carcinogenesis is supported by the genometastatic hypothesis of the phenomenon of tumor-derived circulating cfDNA horizontal transfer into normal cells.51, 52, 53 From this point of view, the presence of large quantities of tumor-derived cfDNAs, including SINEs and LINEs, in the blood of cancer patients, suggests they may carry certain “tumor-induced” properties to normal cells.51
There is some evidence that cfDNA can be transferred horizontally in recipient cells by apoptotic bodies, virosomes, and exosomes.53, 54, 55 Our data show that murine-specific SINEs and LINEs, derived from blood of LLC-bearing mice, can be captured in naked form by cells of human origin, evidence of the ability of tandem repeats to participate in interspecies horizontal gene transfer. The anti-metastatic effect of DNase I may be explained with the hypothesis of genometastases: DNase I causes a degradation of cfDNAs, including SINEs and LINEs, thus reducing the rate of metastases spreading.
Thus, DNase I reduced the levels of SINEs and LINEs in the bloodstream of tumor-bearing mice, which correlated with a pronounced anti-metastatic effect on several murine tumor models. To conclude, we can postulate that tandem repeats related to SINE and LINE retrotransposons can be used as universal markers of carcinogenesis, and by all appearances play important roles in metastases development.
Materials and Methods
Cell Cultures
Human epidermoid carcinoma KB-3-1 cell line was purchased from the Institute of Cytology (Russian Academy of Sciences, St. Petersburg, Russia). Mouse melanoma B16 cell line was purchased from N. N. Blokhin Cancer Research Center, Moscow, Russia. The modified mouse MDR-positive lymphosarcoma RLS40 cells were obtained from the cell collection of the Institute of Chemical Biology and Fundamental Medicine (SB RAS). Mouse Lewis lung carcinoma cell line was generously provided by Dr. N.A. Popova (Institute of Cytology and Genetics, SB RAS). B16 and KB-3-1 cells were grown on DMEM, containing 10% fetal bovine serum (FBS) and 1% antibiotic-antimycotic solution (10 mg/mL streptomycin, 10,000 IU/mL penicillin, and 25 μg/mL amphotericin (IMP Biomedicals, Germany) at 37°C in a humidified atmosphere with 5% CO2. RLS40 and LLC cells were grown on IMDM media, containing 10% FBS, 100 units/mL penicillin, 100 μg/mL streptomycin, 2 mM glutamine, and 40 mM vinblastine at 37°C, in a humidified atmosphere with 5% CO2.
Tumor Strains
The LLC tumor strain was obtained from the vivarium of The Institute of Cytology and Genetics (ICG) SB RAS, Novosibirsk, Russia. RLS40 are permanently maintained in CBA mice in the form of ascites by intraperitoneal injection of 1 × 106 tumor cells.
Mice
Male 10- to 14-week-old C57BL/6J and CBA/LacSto (hereinafter, C57Bl and CBA) mice were obtained from the vivarium of ICBFM SB RAS, Novosibirsk, Russia. Mice were housed in plastic cages (10 animals per cage) under normal daylight conditions. Water and food were provided ad libitum. All animal procedures were carried out in strict accordance with the approved protocol and recommendations for proper use and care of laboratory animals (ECC Directive 2010/63/EU). The experimental protocols were approved by the Committee on the Ethics of Animal Experiments with the Institute of Cytology and Genetics SB RAS (ethical approval number 22.11 from 30.05.2014) and all efforts were made to minimize suffering.
At the start of the experiments, animal weight (mean ± SD) was 20.2 ± 1.5 g. To perform tumor studies, we used 20 animals per group.
Tumor Transplantation and Design of Animal Experiments
Melanoma B16 Model
To generate a metastatic model of melanoma B16 tumor, we inoculated 1 × 105 cells suspended in 0.2 mL of saline buffer into the lateral tail vein of C57Bl mice. On day 4 after tumor transplantation, mice with B16 were assigned to four groups (n = 20 per group): (1) control, received i.m. saline buffer (0.1 mL), and (2), (3), and (4) received i.m. DNase I at the doses of 0.12, 0.6, and 1.2 mg/kg (0.1 mL).
RLS40 Model
Solid tumors RLS40 were induced by i.m. injection of RLS40 1 × 106 cells suspended in 0.1 mL of saline buffer into the right thighs of CBA mice. On day 4 after tumor transplantation, mice with RLS40 were assigned to three groups (n = 20 per group): (1) control, received i.m. saline buffer (0.1 mL), and (2) and (3) received i.m. DNase I at the doses of 0.12 and 0.24 mg/kg (0.1 mL).
LLC Model
Solid tumors LLC were induced by i.m. injection of LLC 1 × 106 cells suspended in 0.1 mL of saline buffer into the right thighs of C57Bl mice. On day 4 after tumor transplantation, mice with LLC were assigned to two groups (n = 20 per group): (1) control, received i.m. saline buffer (0.1 mL) and (2) received i.m. DNase I at the dose of 0.12 mg/kg (0.1 mL).
In the study, bovine pancreatic DNase I (type IV, lyophilized powder, ≥2,000 Kunitz units/mg protein, Sigma-Aldrich, USA) was used. DNase I was administered daily except for weekends. The total number of injections was 10 for B16 and LLC, and 12 for RLS40.
Tumor size was determined on alternate days with caliper measurements in three perpendicular dimensions. Tumor volumes were calculated as V = (π/6 × length × width × height).
The tumor growth inhibition (TGI) was calculated by the formula: TGI (%) = (V control – V experiment) / V control × 100%, where V control, average tumor volume in the control group (cm3); V experiment, average tumor volume in the DNase I groups (cm3).
On day 22 after B16, on day 15 after LLC, and day 19 after RLS40 transplantation, blood samples (1 mL) were collected from the retro-orbital sinus 60 min after the last injection of DNase I. Then mice were sacrificed and the organs occupied by metastases (lungs and livers) were isolated and fixed in 4% neutral-buffered formaldehyde for subsequent histological analysis.
Analysis of the Number and Area of Metastases
To study anti-metastatic action of DNase I, we carried out evaluation of surface and internal metastases in lungs (melanoma B16 and LLC) and livers (RLS40).
Surface metastases in the lungs (B16 model) were counted using a binocular microscope. Internal metastases in lungs (B16 and LLC models) and livers (RLS40 model) were analyzed using histology. For histological evaluation of internal metastases, fixed lungs and livers were dehydrated in ascending ethanols and xylols and embedded in HISTOMIX paraffin (BioVitrum, Russia). Paraffin sections (5 μm) were sliced on a Microm HM 355S microtome (Thermo Fisher Scientific, USA), stained with hematoxylin and eosin, microscopically examined and scanned. Images were obtained using an Axiostar plus microscope equipped with an Axiocam MRc5 digital camera (Zeiss, Germany). The percentages of the internal metastases areas were determined relative to the total area of sections using Adobe Photoshop Software at magnification ×100.
Inhibition of metastases development was assessed by morphometry using the metastasis inhibition index (MII), calculated as MII = [(mean metastasis areacontrol – mean metastasis areaexperiment) / mean metastasis areacontrol] × 100%. The MII of the control group was taken as 0% and the MII, corresponding to 100%, reflected the absence of metastases.
Each studied group included 20 mice and 10 random fields were studied in each specimen, forming in total 200 fields for each group of mice.
Blood Serum Preparation
Blood samples (1 mL) from tumor-bearing mice treated with saline buffer or DNase I were collected from the retro-orbital sinus 60 min after the last injection. Blood samples from healthy mice (0.2 mL) were collected from the retro-orbital sinus 4 times with an interval of 7 days. Blood serum was prepared from the whole blood by clot formation at 37°C for 30 min and at 4°C overnight, followed by clot discard, and centrifuged (4,000 rpm, 4°C, 20 min) to remove cell debris. Serum samples were stored at −70°C until use.
Isolation of cfDNA from Blood Serum
Serum samples from six animals of each group were pooled according to the groups. cfDNA was isolated from the blood serum by extraction with phenol (phenol-Tris-HCl, pH 8.0) and chloroform, followed by concentration using a QIAquick Gel Extraction Kit (QIAGEN, USA). The concentration of cfDNA was measured by a Qubit fluorometer (Invitrogen, USA) using a Quant-iT dsDNA HS Assay Kit (Invitrogen, USA), according to the manufacturer’s recommendations.
Treatment of KB-3-1 Cells with cfDNA Derived from Blood of LLC-Bearing Mice
KB-3-1 cells were seeded in a 24-well plate (30 × 103 /well) and cultivated in DMEM, supplemented with 10% FBS and 1% antibiotic-antimycotic solution, at 37°С in a humidified atmosphere containing 5% CO2 for 12 h. Medium was discarded and cells were either treated with 100 ng of cfDNAs from the pooled blood serum of healthy or tumor-bearing mice, treated with saline buffer or DNase I (n = 10). After 3 days of incubation, cells were harvested with 0.25% trypsin-EDTA solution (Sigma-Aldrich), washed with PBS, and stored at −20°C before use. The number and viability of harvested cells was measured using Gorjaev’s chamber and Automated Cell Counter TC20 (Bio-Rad, Singapore).
Total DNA Isolation
KB-3-1 cells were washed with PBS twice after harvesting with 0.25% trypsin-EDTA solution (IMP Biomedicals, Germany), mildly trypsinized with 20 μL trypsin, and centrifuged (1,200 rpm) for 20 min at 4°C. Supernatant with surface-bound cfDNA was discarded. Cells were mixed with the lysis buffer (100 mM Tris-HCl, pH 8.0; 5 mM EDTA, pH 8.0; 200 mM NaCl; 0.2% SDS, 10 ng proteinase K) and incubated at 65°C for 4 h. After that DNA was extracted as described above. The concentration of total DNA was measured by a Qubit fluorometer (Invitrogen, USA) using a Quant-iT dsDNA HS Assay Kit (Invitrogen, USA), according to the manufacturer’s recommendations.
Primer Selection
RepeatMasker version 3.1.0 from http://www.repeatmasker.org was used to identify sequences of SINEs and LINEs in the FASTA files from the mouse genome release 31 from Ensembl’s website (http://www.ensembl.org/index.html). The obtained FASTA, containing the sequences of all repeats related to chosen type, was size-selected and clustered using USEARCH free software (http://www.drive5.com/usearch). The obtained consensus sequences were aligned to the mice genome (MM9) using Unipro UGENE 1.26 (http://ugene.net/ru/). Aligned sequences were used for primer selection with IDT PrimerQuest Tool (https://eu.idtdna.com). The list of primers is presented in Table 2.
Real-Time PCR
The alteration of SINEs and LINEs in cfDNAs, from blood plasmа or total DNA from KB-3-1, was detected by SYBR-Green-based quantitative real-time PCR. The PCR mixture (20 μL) consisted of 5 μL of cfDNA (0.1–0.5 ng per reaction), 10 μL of SYBR–green-containing BioMasterCor HS-qPCR (BiolabMix, Russia) and 0.6 μmol of each primer (Table 2). The cycling conditions were as follows: 95°C for 6 min for pre-denaturation; and 95°C for 15 s, 59°C for 20 s, 65°C for 60 s, for 30 cycles; followed by melting curves analysis. The expression level of each gene was indicated by the number of cycles needed for the DNA amplification to reach a threshold. The amount of DNA was calculated from the number of cycles by using standard curves and calculated automatically related to the same healthy sample. The data were analyzed using ΔCT method.
Statistics
All experiments were reproduced in triplicate. Data on inhibition of primary tumor growth were statistically processed using Student’s t test (two tailed, unpaired). MII, metastasis number, and PCR data were statistically processed using one-way ANOVA. Post hoc testing was completed using a post hoc Tukey test; p < 0.05 was considered to be statistically significant. The statistical package STATISTICA version 10.0 was used for analysis.
Author Contributions
L.A.A., A.V.S., and N.L.M. performed all experiments. N.L.M. and M.A.Z. conceived the project and designed the experiments. L.A.A., A.V.S., and N.L.M. participated in critical analysis of the data. All authors contributed to writing of the manuscript.
Acknowledgments
This work was funded by Russian Science Foundation (Grant 19-74-30011) and the Russian State funded budget project of ICBFM SB RAS АААА-А17-117020210024-8. We thank Mrs. Albina V. Vladimirova (ICBFM of SB RAS) for cell maintenance and Dr. Evgenyi V. Brenner for his assistance in data analysis.
References
- 1.Soliman S.E., Alhanafy A.M., Habib M.S.E., Hagag M., Ibrahem R.A.L. Serum circulating cell free DNA as potential diagnostic and prognostic biomarker in non small cell lung cancer. Biochem. Biophys. Rep. 2018;15:45–51. doi: 10.1016/j.bbrep.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rouvinov K., Mermershtain W., Dresler H., Ariad S., Riff R., Shani-Shrem N., Keizman D., Neulander E.Z., Douvdevani A. Circulating cell-free DNA levels in patients with metastatic renal cell carcinoma. Oncol. Res. Treat. 2017;40:707–710. doi: 10.1159/000479523. [DOI] [PubMed] [Google Scholar]
- 3.Tang Z., Li L., Shen L., Shen X., Ju S., Cong H. Diagnostic value of serum concentration and integrity of circulating cell-free DNA in breast cancer: a comparative study with CEA and CA15-3. Lab. Med. 2018;49:323–328. doi: 10.1093/labmed/lmy019. [DOI] [PubMed] [Google Scholar]
- 4.Spindler K.G. Methodological, biological and clinical aspects of circulating free DNA in metastatic colorectal cancer. Acta Oncol. 2017;56:7–16. doi: 10.1080/0284186X.2016.1253861. [DOI] [PubMed] [Google Scholar]
- 5.Haber D.A., Velculescu V.E. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014;4:650–661. doi: 10.1158/2159-8290.CD-13-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang R., Pu W., Zhang S., Chen L., Zhu W., Xiao L., Xing C., Li K. Clinical value of ALU concentration and integrity index for the early diagnosis of ovarian cancer: A retrospective cohort trial. PLoS ONE. 2018;13:e0191756. doi: 10.1371/journal.pone.0191756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sobhani N., Generali D., Zanconati F., Bortul M., Scaggiante B. Cell-free DNA integrity for the monitoring of breast cancer: Future perspectives? World J. Clin. Oncol. 2018;9:26–32. doi: 10.5306/wjco.v9.i2.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leng S., Zheng J., Jin Y., Zhang H., Zhu Y., Wu J., Xu Y., Zhang P. Plasma cell-free DNA level and its integrity as biomarkers to distinguish non-small cell lung cancer from tuberculosis. Clin. Chim. Acta. 2018;477:160–165. doi: 10.1016/j.cca.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Bhargava P.M., Shanmugam G. Uptake of nonviral nucleic acids by mammalian cells. Prog. Nucleic Acid Res. Mol. Biol. 1971;11:103–192. doi: 10.1016/s0079-6603(08)60327-x. [DOI] [PubMed] [Google Scholar]
- 10.Batzer M.A., Deininger P.L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 11.Jelinek W.R., Schmid C.W. Repetitive sequences in eukaryotic DNA and their expression. Annu. Rev. Biochem. 1982;51:813–844. doi: 10.1146/annurev.bi.51.070182.004121. [DOI] [PubMed] [Google Scholar]
- 12.López-Flores I., Garrido-Ramos M.A. The repetitive DNA content of eukaryotic genomes. Genome Dyn. 2012;7:1–28. doi: 10.1159/000337118. [DOI] [PubMed] [Google Scholar]
- 13.Umylny B., Presting G., Efird J.T., Klimovitsky B.I., Ward W.S. Most human Alu and murine B1 repeats are unique. J. Cell. Biochem. 2007;102:110–121. doi: 10.1002/jcb.21278. [DOI] [PubMed] [Google Scholar]
- 14.Adams J.W., Kaufman R.E., Kretschmer P.J., Harrison M., Nienhuis A.W. A family of long reiterated DNA sequences, one copy of which is next to the human beta globin gene. Nucleic Acids Res. 1980;8:6113–6128. doi: 10.1093/nar/8.24.6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stroun M., Anker P., Maurice P., Gahan P.B. Circulating nucleic acids in higher organisms. Int. Rev. Cytol. 1977;51:1–48. doi: 10.1016/s0074-7696(08)60225-9. [DOI] [PubMed] [Google Scholar]
- 16.Cheng J., Cuk K., Heil J., Golatta M., Schott S., Sohn C., Schneeweiss A., Burwinkel B., Surowy H. Cell-free circulating DNA integrity is an independent predictor of impending breast cancer recurrence. Oncotarget. 2017;8:54537–54547. doi: 10.18632/oncotarget.17384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Servomaa K., Rytömaa T. UV light and ionizing radiations cause programmed death of rat chloroleukaemia cells by inducing retropositions of a mobile DNA element (L1Rn) Int. J. Radiat. Biol. 1990;57:331–343. doi: 10.1080/09553009014552441. [DOI] [PubMed] [Google Scholar]
- 18.Iskow R.C., McCabe M.T., Mills R.E., Torene S., Pittard W.S., Neuwald A.F., Van Meir E.G., Vertino P.M., Devine S.E. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee E., Iskow R., Yang L., Gokcumen O., Haseley P., Luquette L.J., 3rd, Lohr J.G., Harris C.C., Ding L., Wilson R.K., Cancer Genome Atlas Research Network Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solyom S., Ewing A.D., Rahrmann E.P., Doucet T., Nelson H.H., Burns M.B., Harris R.S., Sigmon D.F., Casella A., Erlanger B. Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 2012;22:2328–2338. doi: 10.1101/gr.145235.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gualtieri A., Andreola F., Sciamanna I., Sinibaldi-Vallebona P., Serafino A., Spadafora C. Increased expression and copy number amplification of LINE-1 and SINE B1 retrotransposable elements in murine mammary carcinoma progression. Oncotarget. 2013;4:1882–1893. doi: 10.18632/oncotarget.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehner J., Stötzer O.J., Fersching D.M., Nagel D., Holdenrieder S. Plasma DNA integrity indicates response to neoadjuvant chemotherapy in patients with locally confined breast cancer. Int. J. Clin. Pharmacol. Ther. 2013;51:59–62. doi: 10.5414/cpp51059. [DOI] [PubMed] [Google Scholar]
- 23.Oricchio E., Sciamanna I., Beraldi R., Tolstonog G.V., Schumann G.G., Spadafora C. Distinct roles for LINE-1 and HERV-K retroelements in cell proliferation, differentiation and tumor progression. Oncogene. 2007;26:4226–4233. doi: 10.1038/sj.onc.1210214. [DOI] [PubMed] [Google Scholar]
- 24.Helman E., Lawrence M.S., Stewart C., Sougnez C., Getz G., Meyerson M. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 2014;24:1053–1063. doi: 10.1101/gr.163659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodríguez-Martín C., Cidre F., Fernández-Teijeiro A., Gómez-Mariano G., de la Vega L., Ramos P., Zaballos Á., Monzón S., Alonso J. Familial retinoblastoma due to intronic LINE-1 insertion causes aberrant and noncanonical mRNA splicing of the RB1 gene. J. Hum. Genet. 2016;61:463–466. doi: 10.1038/jhg.2015.173. [DOI] [PubMed] [Google Scholar]
- 26.Roberts S.A., Lawrence M.S., Klimczak L.J., Grimm S.A., Fargo D., Stojanov P., Kiezun A., Kryukov G.V., Carter S.L., Saksena G. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stacey S.N., Kehr B., Gudmundsson J., Zink F., Jonasdottir A., Gudjonsson S.A., Sigurdsson A., Halldorsson B.V., Agnarsson B.A., Benediktsdottir K.R. Insertion of an SVA-E retrotransposon into the CASP8 gene is associated with protection against prostate cancer. Hum. Mol. Genet. 2016;25:1008–1018. doi: 10.1093/hmg/ddv622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hancks D.C., Kazazian H.H., Jr. SVA retrotransposons: Evolution and genetic instability. Semin. Cancer Biol. 2010;20:234–245. doi: 10.1016/j.semcancer.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belancio V.P., Roy-Engel A.M., Deininger P.L. All y’all need to know 'bout retroelements in cancer. Semin. Cancer Biol. 2010;20:200–210. doi: 10.1016/j.semcancer.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salganik R.I., Martynova R.P., Matienko N.A., Ronichevskaya G.M. Effect of deoxyribonuclease on the course of lymphatic leukaemia in AKR mice. Nature. 1967;214:100–102. doi: 10.1038/214100a0. [DOI] [PubMed] [Google Scholar]
- 31.Sugihara S., Yamamoto T., Tanaka H., Kambara T., Hiraoka T., Miyauchi Y. Deoxyribonuclease treatment prevents blood-borne liver metastasis of cutaneously transplanted tumour cells in mice. Br. J. Cancer. 1993;67:66–70. doi: 10.1038/bjc.1993.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wen F., Shen A., Choi A., Gerner E.W., Shi J. Extracellular DNA in pancreatic cancer promotes cell invasion and metastasis. Cancer Res. 2013;73:4256–4266. doi: 10.1158/0008-5472.CAN-12-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexeeva L.A., Patutina O.A., Sen’kova A.V., Zenkova M.A., Mironova N.L. Inhibition of invasive properties of murine melanoma by bovine pancreatic DNase I in vitro and in vivo. Mol. Biol. (Mosk.) 2017;51:637–646. doi: 10.7868/S0026898417040024. [DOI] [PubMed] [Google Scholar]
- 34.Patutina O., Mironova N., Ryabchikova E., Popova N., Nikolin V., Kaledin V., Vlassov V., Zenkova M. Inhibition of metastasis development by daily administration of ultralow doses of RNase A and DNase I. Biochimie. 2011;93:689–696. doi: 10.1016/j.biochi.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 35.Alekseeva L.A., Mironova N.L., Brenner E.V., Kurilshikov A.M., Patutina O.A., Zenkova M.A. Alteration of the exDNA profile in blood serum of LLC-bearing mice under the decrease of tumour invasion potential by bovine pancreatic DNase I treatment. PLoS ONE. 2017;12:e0171988. doi: 10.1371/journal.pone.0171988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patutina O.A., Mironova N.L., Ryabchikova E.I., Popova N.A., Nikolin V.P., Kaledin V.I., Vlassov V.V., Zenkova M.A. Tumoricidal Activity of RNase A and DNase I. Acta Naturae. 2010;2:88–94. [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y., Cai H., Yuan X., Xu H., Hu Y., Rui X., Wu J., Chen J., Li J., Gao X., Yin D. Efficient targeting drug delivery system for Lewis lung carcinoma, leading to histomorphological abnormalities restoration, physiological and psychological statuses improvement, and metastasis inhibition. Mol. Pharm. 2018;15:2007–2016. doi: 10.1021/acs.molpharmaceut.8b00161. [DOI] [PubMed] [Google Scholar]
- 38.Mironova N.L., Petrushanko I.Y., Patutina O.A., Sen’kova A.V., Simonenko O.V., Mitkevich V.A., Markov O.V., Zenkova M.A., Makarov A.A. Ribonuclease binase inhibits primary tumor growth and metastases via apoptosis induction in tumor cells. Cell Cycle. 2013;12:2120–2131. doi: 10.4161/cc.25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giavazzi R., Decio A. Syngeneic murine metastasis models: B16 melanoma. Methods Mol. Biol. 2014;1070:131–140. doi: 10.1007/978-1-4614-8244-4_10. [DOI] [PubMed] [Google Scholar]
- 40.Park J., Wysocki R.W., Amoozgar Z., Maiorino L., Fein M.R., Jorns J., Schott A.F., Kinugasa-Katayama Y., Lee Y., Won N.H. Cancer cells induce metastasis-supporting neutrophil extracellular DNA traps. Sci. Transl. Med. 2016;8:361ra138. doi: 10.1126/scitranslmed.aag1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trejo-Becerril C., Pérez-Cardenas E., Gutiérrez-Díaz B., De La Cruz-Sigüenza D., Taja-Chayeb L., González-Ballesteros M., García-López P., Chanona J., Dueñas-González A. Antitumor Effects of Systemic DNAse I and Proteases in an In Vivo Model. Integr. Cancer Ther. 2016;15:NP35–NP43. doi: 10.1177/1534735416631102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alcázar-Leyva S., Cerón E., Masso F., Montaño L.F., Gorocica P., Alvarado-Vásquez N. Incubation with DNase I inhibits tumor cell Proliferation. Med. Sci. Monit. 2009;15:CR51–CR55. [PubMed] [Google Scholar]
- 43.Hussein N.A., Mohamed S.N., Ahmed M.A. Plasma ALU-247, ALU-115, and cfDNA integrity as diagnostic and prognostic biomarkers for breast cancer. Appl. Biochem. Biotechnol. 2019;187:1028–1045. doi: 10.1007/s12010-018-2858-4. [DOI] [PubMed] [Google Scholar]
- 44.Sciamanna I., Landriscina M., Pittoggi C., Quirino M., Mearelli C., Beraldi R., Mattei E., Serafino A., Cassano A., Sinibaldi-Vallebona P. Inhibition of endogenous reverse transcriptase antagonizes human tumor growth. Oncogene. 2005;24:3923–3931. doi: 10.1038/sj.onc.1208562. [DOI] [PubMed] [Google Scholar]
- 45.Mangiacasale R., Pittoggi C., Sciamanna I., Careddu A., Mattei E., Lorenzini R., Travaglini L., Landriscina M., Barone C., Nervi C. Exposure of normal and transformed cells to nevirapine, a reverse transcriptase inhibitor, reduces cell growth and promotes differentiation. Oncogene. 2003;22:2750–2761. doi: 10.1038/sj.onc.1206354. [DOI] [PubMed] [Google Scholar]
- 46.Stroun M., Lyautey J., Lederrey C., Mulcahy H.E., Anker P. Alu repeat sequences are present in increased proportions compared to a unique gene in plasma/serum DNA: evidence for a preferential release from viable cells? Ann. N Y Acad. Sci. 2001;945:258–264. doi: 10.1111/j.1749-6632.2001.tb03894.x. [DOI] [PubMed] [Google Scholar]
- 47.Xue X., Zhu Y.M., Woll P.J. Circulating DNA and lung cancer. Ann. N Y Acad. Sci. 2006;1075:154–164. doi: 10.1196/annals.1368.021. [DOI] [PubMed] [Google Scholar]
- 48.Holmes S.E., Dombroski B.A., Krebs C.M., Boehm C.D., Kazazian H.H., Jr. A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat. Genet. 1994;7:143–148. doi: 10.1038/ng0694-143. [DOI] [PubMed] [Google Scholar]
- 49.Katzir N., Rechavi G., Cohen J.B., Unger T., Simoni F., Segal S., Cohen D., Givol D. “Retroposon” insertion into the cellular oncogene c-myc in canine transmissible venereal tumor. Proc. Natl. Acad. Sci. USA. 1985;82:1054–1058. doi: 10.1073/pnas.82.4.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kingsmore S.F., Giros B., Suh D., Bieniarz M., Caron M.G., Seldin M.F. Glycine receptor beta-subunit gene mutation in spastic mouse associated with LINE-1 element insertion. Nat. Genet. 1994;7:136–141. doi: 10.1038/ng0694-136. [DOI] [PubMed] [Google Scholar]
- 51.García-Olmo D.C., Domínguez C., García-Arranz M., Anker P., Stroun M., García-Verdugo J.M., García-Olmo D. Cell-free nucleic acids circulating in the plasma of colorectal cancer patients induce the oncogenic transformation of susceptible cultured cells. Cancer Res. 2010;70:560–567. doi: 10.1158/0008-5472.CAN-09-3513. [DOI] [PubMed] [Google Scholar]
- 52.Bergsmedh A., Szeles A., Henriksson M., Bratt A., Folkman M.J., Spetz A.L., Holmgren L. Horizontal transfer of oncogenes by uptake of apoptotic bodies. Proc. Natl. Acad. Sci. USA. 2001;98:6407–6411. doi: 10.1073/pnas.101129998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaiffe E., Prétet J.L., Launay S., Jacquin E., Saunier M., Hetzel G., Oudet P., Mougin C. Apoptotic HPV positive cancer cells exhibit transforming properties. PLoS ONE. 2012;7:e36766. doi: 10.1371/journal.pone.0036766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gahan P.B., Stroun M. The virtosome-a novel cytosolic informative entity and intercellular messenger. Cell Biochem. Funct. 2010;28:529–538. doi: 10.1002/cbf.1690. [DOI] [PubMed] [Google Scholar]
- 55.García-Olmo D., García-Olmo D.C., Ontañón J., Martinez E. Horizontal transfer of DNA and the “genometastasis hypothesis”. Blood. 2000;95:724–725. [PubMed] [Google Scholar]