Abstract
The aim of this work was to verify the occurrence, quantification, pulse types, and antimicrobial susceptibility profiles of Salmonella sp. isolated from chicken meat produced and marketed in the state of Paraná, considered to be the state with the highest production of poultry meat in Brazil. Ninety-five of 300 (31.5%) frozen cuts of chicken were found to contain Salmonella sp., and 98 different isolates of Salmonella sp. were cultured from the positive samples. Quantification showed low Salmonella sp. loading, ranging from 0.12 to 6.4 MPN/g. The antimicrobial resistance test was performed against 16 agents from 6 different classes. All isolates were sensitive to meropenem, imipenem, chloramphenicol, and amikacin. The highest resistance rates were observed for nalidixic acid (95%), tetracycline (94%), doxycycline (94%), ampicillin (87%), amoxicillin with clavulanic acid (84%), ceftriaxone (79%), and ciprofloxacin (76%). A total of 84 (85.7%) of the isolates were identified with a multidrug resistant profile, 13 of which were found to have encoding genes extended-spectrum beta-lactamase (ESBL), especially blaCTX-M-2 e blaTEM-1. The major serovars identified were S. Typhimurium (43%) and S. Heidelberg (39%). The third most isolated serovar was S. Ndolo (6%), without previous reports of its presence in poultry meat in Brazil. Molecular characterization of S. Typhimurium and S. Heidelberg isolates by pulsed field gel electrophoresis (PFGE) showed a clonal relationship between all isolates of the same serovar (genetic similarity greater than 80%). Isolates of S. Typhimurium and S. Heidelberg with 100% similarity were found in up to five different geographic regions of the state, showing the potential for the spread of this pathogen in the Paraná poultry chain. Epidemiological surveys like this are important to understand the dynamics of dissemination and to monitor the prevalence of pathogens in the final products of poultry chains. In addition, to know the resistance profile of strains of Salmonella sp. present in food that contributes to the adoption of faster and more effective therapeutic measures, when necessary.
Keywords: blaCTX-M-2, blaTEM-1, PFGE, Sequencing, Salmonella Ndolo, mMPN
Introduction
Salmonella sp. is one of the main agents involved in outbreaks of foodborne illnesses in Brazil, Europe, and USA [1–3], with non-typhoid Salmonella accounting for about 94 million of cases of gastroenteritis, causing 155,000 deaths per year, worldwide [4]. The intensity of clinical symptoms depends on factors related to the microorganism, such as the serovar and bacterial load ingested, and also on factors related to the individual, such as age, decreased gastric acidity and intestinal motility, changes in the intestinal microbiota, diabetes mellitus, inflammatory diseases, and alterations in the function of macrophages [5, 6]. In most patients with salmonellosis, spontaneous resolution of the disease occurs without clinical or drug intervention. However, in some individuals, there may be clinical worsening, with evolution to bacteremia, meningitis, or severe diarrhea, which requires antibiotic therapy [7]. The list of therapeutic options for these patients is becoming increasingly reduced, since the expression of ESBL enzymes associated with other mechanisms is responsible for the resistance of Salmonella sp. to certain drugs [8].
Between 2000 and 2017, 12,503 foodborne outbreaks were reported in Brazil, resulting in 182 deaths. Among the microorganisms that were characterized, Salmonella sp. appeared as the main agent, reported in approximately 30% of the outbreaks [1]. Chicken meat is one of the main foods involved in the propagation of Salmonella to humans and plays an important role in the distribution of several serovars, since the production chain, ranging from raising of poultry to culinary preparation, allows the meat to be contaminated at all stages [9–15].
Brazil is the largest exporter and the second largest producer of chicken meat in the world. The state of Paraná, located in the south of the country, is considered to be the largest Brazilian chicken producer, accounting for just over a third of the national production and exporting its products to more than 150 countries [16, 17]. Despite the productive prominence of the state of Paraná and the relevance of Salmonella sp. to the poultry production chain and the food industry, there are no studies reporting epidemiological data on Salmonella covering all the slaughtering industries in Paraná.
Considering that continuous monitoring of the occurrence and antimicrobial resistance of Salmonella in food is necessary due to the implications of this pathogen for public health and the potential for dissemination of antimicrobial resistant isolates, the aim of the present study was to evaluate the occurrence of Salmonella in chicken cuts produced throughout the territory of Paraná and characterize isolated strains of Salmonella sp. to determine the distribution of serovars, pulse types, and resistance to antimicrobials.
Material and methods
Sample collection and bacterial isolation
Between August 2015 and February 2016, a total of 300 samples of frozen chicken cuts (wing, breast, leg, and fried chicken) were collected from the retail trade of the state of Paraná and the 35 facilities under federal inspection that operate in that state. The total sampling was distributed among all slaughterhouses, resulting in approximately nine samples from each one. The presence of Salmonella was assessed by the ISO 6579:2007 methodology, and pathogen quantification was made by the ISO 6579-2:2012 methodology [18, 19]. As a result of low quantification efficiency observed in the first samples, modifications were made to increase the detection power of the test. Briefly, after weighing the sample (32.5 g) and dilution in 292.5 mL of buffered peptone water BPW, three aliquots of 25 mL were transferred to a series of three tubes, and 7.5 mL was divided into three wells in a 24-well cell culture dish. The remainder of the sample volume was used for determination of the presence/absence of Salmonella sp. Then, 500 μL of each of the first wells were transferred to subsequent wells, which contained 2 mL of BPW. The procedure was repeated in three further seeding sequences until four dilutions were obtained. Each 24-well plate was divided into two, making it possible to perform the mMPN simultaneously from two different samples. The tubes and the 24-well plate were incubated along with the remainder of the sample volume. Selective enrichment was carried out on Rappaport-Vassiliadis semi-solid agar (MRSV) and selective plating on xylose lysine deoxycholate agar (XLD) from each well and the tubes, with typical colonies being tested biochemically. Characteristic Salmonella isolates were purified and stored at − 20 °C in duplicate for further testing.
The numerical results of the mMPN technique were obtained with the aid of the MPN calculation program [20], version 3 and expressed in MPN/g of the sample.
Confirmation of the Salmonella genus
The suspected Salmonella isolates were confirmed by PCR through the identification of the invA gene. Extraction of bacterial DNA from the isolates was carried out with the Wizard® Genomic DNA Purification kit (Promega Madison, Wisconsin, USA), starting from cultures resuspended in BHI broth (brain heart infusion), after 24 hours of incubation at 35–37 °C. Amplification followed the protocol described by Swamy et al. [21], using a strain of Salmonella Typhimurium ATTCC® 14028 as positive control and a sample of ultrapure water as a negative control.
Serotyping
Confirmed Salmonella isolates were serotyped based on the somatic (O) and flagellar (H) antigens by the enterobacteria laboratory of the Oswaldo Cruz Foundation, located in the state of Rio de Janeiro, Brazil.
Antimicrobial susceptibility test and ESBL production
Antimicrobial susceptibility testing and interpretation were performed according to the M31-A3 [22] and M100-S23 [23] standards of the Clinical Laboratory Standards Institute by the agar-diffusion method using the strain Escherichia coli ATCC® 25922 as a control.
The following antimicrobial agents were tested: ampicillin (10 μg), cefepime (30 μg), ceftriaxone (30 μg), meropenem (10 μg), imipenem (10 μg), aztreonam (30 μg), amoxicillin with clavulanic acid (30 μg), doxycycline (30 μg), nalidixic acid (30 μg), ciprofloxacin (5 μg), sulfamethoxazole with trimethoprim (10 μg), gentamicin (10 μg), amikacin (30 μg), tobramycin (23.75/1.25 μg), and chloramphenicol (30 μg).
The isolates that showed resistance to at least one beta-lactam agent were tested for production of ESBL enzymes by the dual disc diffusion method [24]. Escherichia coli ATCC® 25922 was used as a control. Presence of blaCTX-M, blaOXA, blaSHV, ampC, and blaTEM genes in positive isolates was determined by PCR according to protocols described by Belaaouaj et al. [25], M’zali et al. [26], Féria [27], and Edelstein et al. [28], respectively.
The amplified genes were purified with the QIAquick® PCR Purification Kit (Qiagen Inc., Valencia, CA, USA) and sent to the Human Genome and Stem Cell Research Center of the Institute of Biosciences of the University of São Paulo (USP), Brazil, for sequencing. The sequences obtained were concatenated using Mega 7 software and then analyzed on GenBank, using the online BLAST tools.
Pulsed field gel electrophoresis
Isolates of Salmonella sp. were identified by using the pulsed field gel electrophoresis (PFGE), according to the protocol described by Ribot et al. [29]. Briefly, proteinase K was used for cell lysis and the XbaI enzyme for DNA digestion. The DNA fragments were separated in a CHEF-DRIII apparatus (Bio-Rad Laboratories, Hercules, CA, USA) and standardized with a strain of Salmonella enterica subsp. enterica serovar Braenderup ATCC® BAA-664TM.
The images obtained were analyzed using the BioNumerics software version 7.6 (Applied-Maths, Kortrijk, Belgium) and the band patterns were compared using the Dice similarity coefficient with a tolerance of 1.5% and UPGMA (unweighed pair group method using arithmetic average), considering clonally related isolates with similarity greater than 80%.
Statistical analysis
A 4 by 2 contingency table and the chi-square test were used to determine if there was a difference between the occurrences of Salmonella sp. in different types of chicken cuts. Values of p < 0.05 were considered significant. The analysis was performed using the BioEstat 5.3 program.
Results
Quantification and identification de Salmonella sp.
The most probable number test of Salmonella sp. per gram of product presented quantifiable results in only 7.7% (23/300) of the samples analyzed. The values of Salmonella sp. counts ranged from 0.12 to 6.4 MPN/g of analyzed product, with most of the samples having values lower than 3 MPN/g (Table 1). The two samples with higher counts had values of 3.1 and 6.4 MPN/g.
Table 1.
Distribution of chicken cut samples that showed quantifiable mMPN values of Salmonella sp.
| MPN/g | Number of samples | % |
|---|---|---|
| 0.030–0.30 | 12 | 52 |
| 0.301–3.00 | 9 | 39 |
| 3.01–30.00 | 2 | 9 |
| TOTAL | 23 | 100 |
Salmonella sp. were present in 30% (90/300) of the samples analyzed. Five other samples were negative in the qualitative analysis but showed Salmonella sp. by mMPN. Thus, 95 (31.7%) samples of chicken cuts produced in the state of Paraná were considered positive for Salmonella sp. (Table 2). Of these 95 samples, 98 isolates were selected, and all were confirmed as Salmonella sp. by PCR detection of the invA gene.
Table 2.
Occurrence and quantification of Salmonella sp. in frozen chicken cuts produced in the region and state of Paraná
| Region | Number of poultry slaughterhouses | Samples with Salmonella sp./analyzed samples | Samples with Salmonella sp. (%) | Number of samples with Salmonella spp. quantifiable | MPN of Salmonella sp./gram of product | |
|---|---|---|---|---|---|---|
| Average | Minimum–maximum | |||||
| Central north | 12 | 28/102 | 27.5 | 10 | 0.85 | 0.12–6.4 |
| West | 7 | 24/60 | 40 | 1 | 2 | 2–2 |
| Northwest | 5 | 21/43 | 48.8 | 7 | 0.74 | 0.14–3.1 |
| Southwest | 5 | 8/43 | 18.6 | 1 | 0.14 | 0.14–0.14 |
| Pioneer north | 2 | 3/17 | 17.6 | 0 | ND | ND |
| Western center | 2 | 7/17 | 41.2 | 1 | 0.58 | 0.58–0.58 |
| Eastern center | 1 | 4/9 | 44.4 | 3 | 0.7 | 0.14–1.8 |
| Metropolitan | 1 | 0/9 | 0 | 0 | - | - |
| Total | 35 | 95/300 | 31.7 | 23 | ||
ND, not detectable by the mMPN technique (< 1 MPN/g)
The chi-square test showed that the occurrence of Salmonella sp. did not differ between the different chicken cuts (p > 0.05) (Table 3).
Table 3.
Distribution of chicken samples positive for Salmonella sp., considering the type of cut analyzed
| Type of cut | Samples with Salmonella sp./analyzed samples | Samples with Salmonella sp. (%) |
|---|---|---|
| Winga | 28/71 | 39.4* |
| Fried chicken | 17/54 | 31.5* |
| Breastb | 33/103 | 32* |
| Legc | 17/72 | 23.6* |
| Total | 300 | - |
aMiddle of wing, drumstick, and whole wing
bBreast filet, filet, and whole breast
cChicken thigh, chicken upper leg, and whole leg
*Indicates that there was no statistical difference in the comparison between the number of positive samples by the chi-square test at 5% probability
Nine different serovars were identified among the 98 isolates of Salmonella sp. (Table 4). In the three samples where more than one serovar was identified, associations of Salmonella Heidelberg with Salmonella Ndolo, Salmonella Typhimurium with Salmonella Ndolo, and Salmonella Heidelberg with Salmonella Typhimurium were found.
Table 4.
Serovars of Salmonella sp. found in chicken cuts produced in the state of Paraná between 2015 and 2016
| Serovar | Number of isolates/total of isolates | % |
|---|---|---|
| Salmonella Typhimurium | 42/98 | 43 |
| Salmonella Heidelberg | 38/98 | 39 |
| Salmonella Ndolo | 6/98 | 6 |
| Salmonella Minnesota | 4/98 | 4 |
| Salmonella enterica subsp. enterica (O:4,5) | 2/98 | 2 |
| Salmonella Thompson | 2/98 | 2 |
| Salmonella Schwarzengrund | 2/98 | 2 |
| Salmonella enterica subsp. enterica (O:3,10:e,h) | 1/98 | 1 |
| Salmonella Abony | 1/98 | 1 |
Antimicrobial resistance profile
All isolates were sensitive to meropenem, imipenem, chloramphenicol, and amikacin. Only 5.1% (5/98) of the isolates were sensitive to all antimicrobials tested. The remaining isolates were resistant to two or more classes of antimicrobials (Table 5). The highest resistance rates were observed for nalidixic acid (95%), tetracycline (94%), doxycycline (94%), ampicillin (87%), amoxicillin with clavulanic acid (84%), ceftriaxone (79%), and ciprofloxacin (76%). In 85.7% (84/98) of the isolates, multi-resistance was observed, that is, resistance to three or more classes of antimicrobials. The most prevalent profile among the multi-resistant isolates was resistance to ampicillin, amoxicillin with clavulanic acid, doxycycline, tetracycline, nalidixic acid, and ceftriaxone, which was observed in 37 isolates (37.8%).
Table 5.
Resistance phenotypes of Salmonella sp. isolates with regard to tested antimicrobial classes
| Number of classes with resistance | Classes of antimicrobials | Number of isolates | % |
|---|---|---|---|
| 0 | Sensitive to all classes of antimicrobials | 05 | 5.1 |
| 1 | - | - | - |
| 2 | Quinolones/tetracyclines | 07 | 7.1 |
| Beta-lactams/quinolones | 02 | 2.0 | |
| 3 | Beta-lactams/quinolones/tetracyclines | 67 | 68.5 |
| Quinolones/aminoglycosides/tetracyclines | 02 | 2.0 | |
| 4 | Beta-lactams/aminoglycosides/tetracyclines/quinolones | 13 | 13.3 |
| Beta-lactams/folate inhibitors/tetracyclines/quinolones | 01 | 1.0 | |
| Beta-lactams/aminoglycosides/tetracyclines/folate inhibitors | 01 | 1.0 | |
| Total | 98 | 100 |
Resistance to at least one of the beta-lactam agents, ceftriaxone or cefepime, was observed in 77 isolates, and these isolates were subjected to phenotypic screening for production of ESBL enzymes. After this screening, 13 isolates with phenotypic patterns compatible with production of these enzymes were submitted to PCR to investigate genes responsible for ESBL expression. The blaCTX-M gene was found in all 13 isolates and association of the blaCTX-M gene with the blaTEM gene was found in three isolates. Sequencing showed these genes to be blaCTX-M-2 and blaTEM-1 in all cases (Table 6).
Table 6.
ESBL enzyme production characteristics of Salmonella sp. isolates obtained from chicken cuts produced in the state of Paraná
| Serovar | Phenotype of resistance | MR | Type of ESBL |
|---|---|---|---|
| Typhimurium | AMP, GEN, DOX, TET, NAL, CRO, CPM, ATM | Yes | CTX-M-2 |
| Heidelberga | AMP, AMC, DOX, TET, NAL, TOB, CRO, CPM, ATM | Yes | CTX-M-2 |
| Ndoloa | AMP, GEN, DOX, TET, CRO, CPM, ATM | Yes | CTX-M-2 |
| Heidelberg | AMP, GEN, DOX, TET, NAL, CRO, CPM, ATM | Yes |
CTX-M-2 TEM-1 |
| Heidelberg | AMP, GEN, DOX, TET, NAL, CRO, CPM, ATM | Yes | CTX-M-2 |
| Ndolo | AMP, GEN, DOX, TET, NAL, CRO, CPM, ATM | Yes | CTX-M-2 |
| Typhimurium | AMP, DOX, TET, NAL, CRO, CPM, ATM | Yes |
CTX-M-2 TEM-1 |
| Typhimurium | AMP, DOX, TET, NAL, CRO, CPM, ATM | Yes |
CTX-M-2 TEM-1 |
| Heidelberg | AMP, GEN, DOX, TET, NAL, CRO, CPM, ATM | Yes | CTX-M-2 |
| Typhimurium | AMP, GEN, DOX,TET, NAL, CRO, CPM, ATM | Yes | CTX-M-2 |
| Typhimurium | AMP, GEN, DOX,TET, NAL, CRO, CPM, ATM | Yes | CTX-M-2 |
| Ndolo | AMP, GEN, DOX, TET, NAL, CRO, CPM, ATM | Yes | CTX-M-2 |
| Typhimurium | AMP, GEN, DOX, TET, NAL, CRO, CPM, ATM | Yes | CTX-M-2 |
MR, multi-resistance; AMP, ampicillin; GEN, gentamicin; DOX, doxycycline; TET, tetracycline; NAL: nalidixic acid; CRO, ceftriaxone; CPM, cefepime; ATM, aztreonam; AMC, amoxicillin with clavulanic acid; TOB, tobramycin
aIsolates from the same sample
PFGE
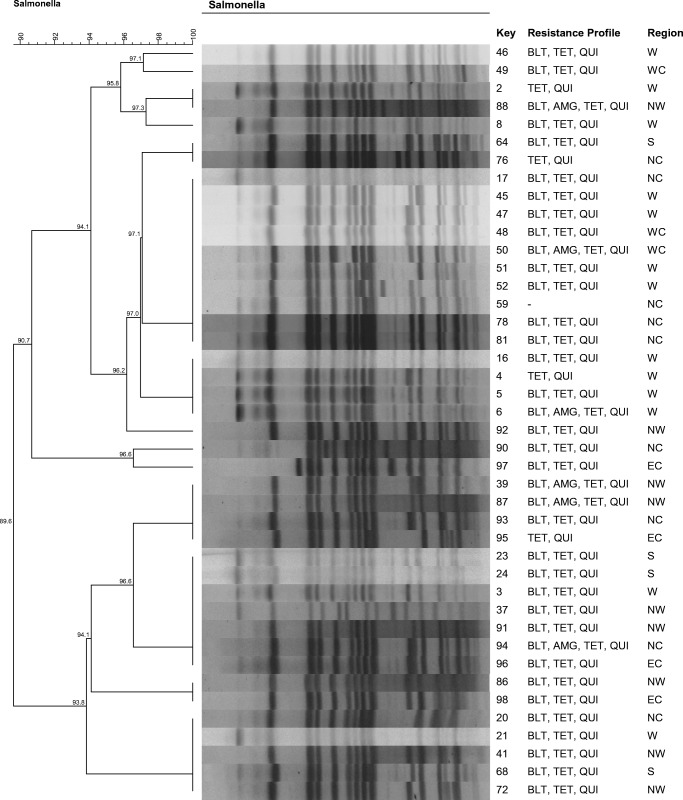
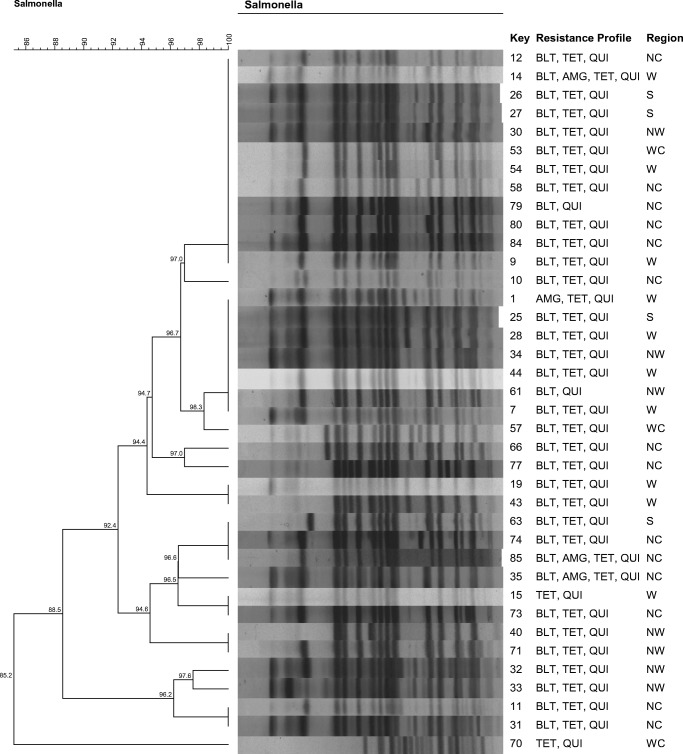
To evaluate genetic similarity, two images were obtained, the first using only Salmonella Typhimurium (42 strains) and the second using only Salmonella Heidelberg (38 strains), considering the predominance of these two serovars among the isolates. The others serovars were not used to the PFGE analysis.
All S. Typhimurium isolates were clonally related (genetic similarity greater than 80%). Isolates from up to five geographic regions (southwest, west, northwest, north central, eastern center) demonstrated 100% genetic similarity (Fig. 1).
Fig. 1.
Pulse types (XbaI) and antimicrobial sensitivity profiles of the 42 Salmonella Typhimurium isolates obtained from chicken cuts produced in the state of Paraná between August 2015 and February 2016. BLT, beta-lactams; TET, tetracyclines; QUI, quinolones; AMG, aminoglycosides; W, west; WC, western center; NW, northwest; S, southwest; NC, north central; EC, eastern center
Similarly, all S. Heidelberg isolates showed are clonally related. Isolates with identical genetic profiles were also identified in five distinct geographic regions (north central, west, southwest, northwest and western center) (Fig. 2).
Fig. 2.
Pulse types (XbaI) and antimicrobial sensitivity profiles of the 38 Salmonella Heidelberg isolates obtained from chicken cuts produced in the state of Paraná between August 2015 and February 2016. BLT, beta-lactams; TET, tetracyclines; QUI, quinolones; AMG, aminoglycosides; NC, north central; W, west; S, southwest; WC, western center; NW, northwest
Discussion
The occurrence of Salmonella sp. determined in the present study can be considered elevated and resembles that determined in other national studies [30, 31]. On the other hand, the contaminant load in these samples was low, based on levels described by Yamatogi et al. [32] and by the American microbiological program for data collection on chicken cuts (RCPBS) [33]. However, human salmonellosis due to the consumption of these chicken cuts should not be neglected, since even this small number of cells could persist and multiply in food when there are temperature abuses during storage or due to incorrect handling and heat treatment [34–36]. Inadequate safety during handling, besides facilitating multiplication of Salmonella sp. initially present in poultry meat, can also transfer these cells to other foods when they are prepared together, mainly in the domestic environment [37]. This cross-contamination becomes more important when the food involved will not be subjected to heat treatment before consumption, representing a great risk in the development of salmonellosis [38].
Samples positive for Salmonella sp. in the mMPN assay but negative in the presence/absence assay indicated heterogeneity in the distribution of Salmonella cells in the evaluated sections, causing pathogen cells to be transferred to the cell culture plate where the counting technique was performed which cannot be detected by the presence/absence technique [39, 40].
Samples positive in the qualitative analysis but negative in the quantitative analysis probably have a number of Salmonella cells below the detection limit of the mMPN test (1 MPN/g), which is lower than the detection limit in the presence/absence technique (0.04 CFU/g) [19]. After addition of more sample volume to the mMPN assay, the samples were able to be quantified, demonstrating that the methodology described by ISO/TS 6579-2 does not present satisfactory results for quantification of Salmonella sp. in poultry meat when the contaminant load is low, as in the present study [33].
The occurrence of Salmonella spp. between the different regions of the state was not statistically compared due to the large variation in the number of samples, determined by the collection methodology. Even so, there is a large variation in the percentage of pathogen occurrence. Salmonella spp. was not identified in any sample of the metropolitan region, suggesting that there may be some effect of environmental temperature on this data, since this region is among the coldest regions of the state [41].
Oscar et al. [42], when evaluating the distribution of contamination by Salmonella spp. in chicken carcasses, they observed that the wings were the most contaminated parts. In this work, no significant difference was observed in the occurrence of Salmonella spp. between the different types of cuts evaluated. Percentages very similar to those found in this study were demonstrated by the RCPBS. This program does not perform the evaluation of fried chicken, but for leg, breast, and wing, the positivity percentages for Salmonella sp. were 24.2%, 27.1%, and 33.3%, respectively, in 2012 [33].
In Brazil, specific sanitary programs to control Salmonella sp. have been in existence since 2003. One of these is the National Program on Poultry Health, which aims to immunize broiler breeders with vaccines against Salmonella Enteritidis, reducing the vertical transmission of the pathogen [43, 44]. It was possible to verify that this serovar was not isolated in any region of the state of Paraná, demonstrating the success of the control measures adopted by the country against serovar Enteritidis; the occurrence of which has been reduced from 84% in the early 2000s to zero, as observed in this study and others [45].
With Salmonella Enteritidis under control, other serovars found a less competitive and more favorable environment to develop, as was likely the case for Salmonella Typhimurium and Salmonella Heidelberg, the main circulating serovars in the state of Paraná, according to the data presented here. In addition to the large percentage of occurrence, the geographical distribution of clonally related isolates (with genetic similarity above 80%) and 100% identical isolates belonging to these two serovars is also notable.
The clonal relationship between all S. Typhimurium isolates and among all S. Heidelberg isolates, as well as the presence of clones of each of these serovars in up to five different regions, suggests that there is circulation of Salmonella sp. strains within the state. This circulation may occur from the purchase of genetic material, raw materials, or other inputs containing Salmonella sp., which may originate from the state itself or from other places. Due to the high production volume and the extent of poultry circulation and inputs from this state, the contamination sources and the dissemination routes of the pathogen in the poultry chain merit more detailed studies.
The serovar Ndolo has only been reported rarely in Brazil. Leal et al. [46] described the presence of this serovar in human isolates from 1978 to 1980, and Hofer et al. [47] reported it in horseflesh slaughtered between 1980 and 1982 in the state of Pernambuco. There have been no previous reports of Salmonella Ndolo in poultry meat in Brazil. In this study, the Ndolo serovar was the third most isolated, demonstrating its emergence in the state of Paraná. Of concern is that half of Salmonella Ndolo isolates had phenotypes related to antibiotic resistance and genes encoding ESBL enzymes. These three isolates were obtained from samples from the northwest and north central regions of Paraná, which are among the three regions with the highest number of slaughterhouses in the state.
High percentages of Salmonella sp. resistant to antimicrobials from the quinolone, tetracycline, and beta-lactam classes have already been reported in Brazil and other countries in isolates of humans and food matrices, showing the global spread of strains resistant to these agents [48–51].
Antimicrobial resistance to quinolones has been steadily increasing. In 1996, resistance to nalidixic acid was reported in 0.4% of Salmonella spp. isolates of poultry meat, in 2008 were already approximately 60% and currently 95% [51, 52]. This resistance has been mainly attributed to mutations in the genes that encode DNA gyrase (gyrA and gyrB) or topoisomerase (parC and parE), preventing the drug from acting on these enzymes [53–55]. However, recently other resistance mechanisms have also been reported, such as plasmid-mediated quinolone resistance (PMQR), which enables chromosomal mutations in target regions of quinolones and reduced susceptibility to this agent [56].
The mechanisms of resistance to tetracyclines undergo constant improvements, as demonstrated by Almeida et al. [57]. The authors identified isolates of Salmonella sp. phenotypically resistant to tetracycline, but without carrying any known resistance gene so far, suggesting the existence of an alternative mode of resistance. This demonstrates the constant need for studies on the subject.
In Brazil, resistance to beta-lactam agents has been mainly associated with CTX-M type ESBL production [58]. The production of these enzymes is usually plasmid-mediated and confers resistance to beta-lactams by hydrolyzing the beta-lactam ring of penicillins, cephalosporins, and related compounds before they reach target binding proteins, inactivating the antibiotic and rendering the therapeutic treatment ineffectively [50, 59–61]. These enzymes are also related to multi-resistant phenotypes, which have already been shown in isolates of Salmonella spp. from poultry slaughter environments in the state of Paraná [62]. The high number of MR isolates shows the need for more rigorous control of the use of antimicrobial agents in animal production in Paraná. Despite the high MR rate, it is necessary to highlight that all isolates were sensitive to carbapenems, since these agents are the first choice to combat microorganisms producing ESBL enzymes [8]. The sensitivity of all isolates to chloramphenicol is a reflection of the ban on the use of this drug in the production of animals in Brazil since 1998 [63]. Since then, the resistance of Salmonella sp. to this antimicrobial has been reported less frequently. In 2003, 27.6% of Salmonella sp. isolates were resistant to chloramphenicol, a percentage which was gradually reduced to zero, as observed in the present study [62, 64, 65]. The lack of chloramphenicol resistance can be explained by the absence of selective pressure, inducing the bacteria to evolve without the need for a resistance gene.
With the increasing development of antimicrobial resistance mechanisms already known, veterinary and human medicine must constantly seek new alternatives for the prevention and treatment of human Salmonella sp. infections. One of the promising novelties in the medical field is the development of mono and divalent vaccines against Salmonella Typhimurium and Salmonella Enteritidis [66]. Tools such as these become fundamental, primarily for patients in at-risk groups for whom failure of antimicrobial protocols can determine the mortality.
The data obtained through this active epidemiological surveillance can contribute to early clinical and microbiological diagnoses and help guide appropriate treatment in patients with salmonellosis. In addition, these data also help to reduce the time required for the adoption of preventive measures in the food industry, making it possible to target actions on the most prevalent serovars in the state. These measures are necessary to achieve more stringent sanitary standards in order to reduce the impact of this pathogen on public health.
Conclusions
Despite all the sanitary measures adopted by the Brazilian poultry system for the control of Salmonella sp. and the low microbial load observed, the occurrence of Salmonella and of antimicrobial resistance in chicken meat marketed in the state of Paraná can still be considered high. The distribution of the 100% identical multi-resistant isolates in several regions demonstrates the movement of the pathogen in the state, indicates an increased risk to food safety, and reinforces the need for constant surveillance of this pathogen. It is necessary to be more prudent in the use of antimicrobials in the poultry production system of Paraná and to adopt more effective Salmonella sp. control measures in poultry breeding and slaughtering establishments, especially those based on risk analyses.
Funding information
This study was funded in part by the Coordination of Superior Level Staff Improvement—Brazil (CAPES)—Finance Code 001, National Council for Scientific and Technological Development (CNPq).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Brazil (2018) Outbreaks of foodborne diseases in Brazil. http://portalarquivos2.saude.gov.br/images/pdf/2018/julho/02/Apresentacao-Surtos-DTA-Junho-2018.pdf. Accessed 18 August 2018
- 2.Marder EP, Griffin PM, Cieslak PR, Dunn J, Hurd S, Jervis R, Lathrop S, Muse A, Ryan P, Smith K, Tobin-D’angelo M, Vugia DJ, Holt KG, Wolpert BJ, Tauxe R, Geissler AL. Preliminary Incidence and trends of infections with pathogens transmitted commonly through food—foodborne diseases active surveillance network, 10 U.S. sites, 2006–2017. MMWR Morb Mort Wkly Rep. 2018;67:324–328. doi: 10.15585/mmwr.mm6711a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.EFSA (European Food Safety Authority) The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J. 2017;15:5077. doi: 10.2903/j.efsa.2017.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majowicz SE, Musto J, Scallan E, Angulo FJ, O’Brien SJ, Jones TF, Fazil A, Hoekstra RM. The global burden of nontyphoidal gastroenteritis. Clin Infect Dis. 2010;50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 5.Abe K, Saito N, Kasuga F, Yamamoto S. Prolonged incubation period of salmonellosis associated with low bacterial doses. J Food Prot. 2004;67:2735–2740. doi: 10.4315/0362-028X-67.12.2735. [DOI] [PubMed] [Google Scholar]
- 6.Vázquez EG, Torres AH, Martínez JAH, Gómez JG. Infecciones por Salmonella y Yersinia. Medicine (Spain) 2014;11:3322–3326. doi: 10.1016/S0304-5412(14)70777-2. [DOI] [Google Scholar]
- 7.Batchelor M, Hopkins KL, Threlfall EJ, Clifton-Hadley FA, Stallwood AD, Davies RH, Liebana E. Characterization of AmpC-mediated resistance in clinical Salmonella isolates recovered from humans during the period 1992 to 2003 in England and Wales. J Clin Microbiol. 2005;43:2261–2265. doi: 10.1128/JCM.43.5.2261-2265.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vardakas KZ, Tansarli GS, Rafailidis PI, Falagas ME. Carbapenems versus alternative antibiotics for the treatment of bacteraemia due to enterobacteriaceae producing extended-spectrum beta-lactamases: a systematic review and meta-analysis. J Antimicrob Chemother. 2012;67:2793–2803. doi: 10.1093/jac/dks301. [DOI] [PubMed] [Google Scholar]
- 9.Djeffal S, Mamache B, Elgroud R, Hireche S, Bouaziz O. Prevalence and risk factors for Salmonella spp. contamination in broiler chicken farms and slaughterhouses in the northeast of Algeria. Vet World. 2018;11:1102–1108. doi: 10.14202/vetworld.2018.1102-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, Fu Y, Xiong Z, Ma Y, Wei Y, Qu X, Zhang H, Zhang J, Liao M. Highly prevalent multidrug-resistant Salmonella from chicken and pork meat at retail markets in Guangdong, China. Front Microbiol. 2018;9:1–9. doi: 10.3389/fmicb.2018.02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gould LH, Walsh K, Vieira AR, Herman K, Williams IT, Hall AJ, Cole D. Surveillance for foodborne disease outbreaks—United States, 1998–2008. MMWR. 2013;62:1–34. [PubMed] [Google Scholar]
- 12.Saeed AA, Hasoon MF, Mohammed MH. Isolation and molecular identification of Salmonella typhimurium from chicken meat in Iraq. J World’s Poult Res. 2013;3:63–67. [Google Scholar]
- 13.El-Aziz DMA. Detection of Salmonella Typhimurium in retail chicken meat and chicken giblets. Asian Pac J Trop Biomed. 2013;3:678–681. doi: 10.1016/S2221-1691(13)60138-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hur J, Jawale C, Lee JH. Antimicrobial resistance of Salmonella isolated from food animals: a review. Food Res Int. 2012;45:819–830. doi: 10.1016/j.foodres.2011.05.014. [DOI] [Google Scholar]
- 15.Lillard HS. The impact of commercial processing procedures on the bacterial contamination and cross-contamination of broiler carcasses. J Food Prot. 1990;53:202–204. doi: 10.4315/0362-028X-53.3.202. [DOI] [PubMed] [Google Scholar]
- 16.ABPA (2017) Brazilian Association of Animal Protein. Annual report 2017. http://abpa-br.com.br/storage/files/3678c_final_abpa_ relatorio_anual*_2016_portugues_web_reduzido.pdf. Accessed 10 November 2017
- 17.Sindiavipar (Union of Poultry Products Industries of the State of Paraná) (2016) Press. PR: Poultry exports grow 19% in the first four months of the year. https://www.sindiavipar.com.br/index.php?modulo = 15& acao = detalhe&cod = 175314. Accessed 01 December 2016.
- 18.ISO (2007) Microbiology of food and animal feeding stuffs—horizontal method for the detection of Salmonella spp.—amendment 1: Annex D: detection of Salmonella spp., in animal faeces and in environmental samples from the primary production stage (ISO 6579:2002/Amd. 1:2007).
- 19.ISO/ST (2012) Microbiology of food and animal feeding stuffs—horizontal method for the detection, enumeration and serotyping of Salmonella—part 2: enumeration by a miniaturized most probable technique (ISO/TS 6579-2: 2012).
- 20.MPN Calculation Program (2013) Versão 3. http://standards.iso.org/iso/ts/6579/-2/. Accessed 18 August 2015.
- 21.Swamy SC, Barnhart HM, Lee MD, Dreesen DW. Virulence determinants invA and spvC in Salmonellae isolated from poultry products, wastewater, and human sources. Appl Environ Microbiol. 1996;62:3768–3771. doi: 10.1128/AEM.62.10.3768-3771.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CLSI (Clinical and Laboratory Standards Institute) Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals (M31-A3) 3. Pennsylvania: Wayne; 2008. [Google Scholar]
- 23.CLSI (Clinical and Laboratory Standards Institute) Performance standards for antimicrobial susceptibility testing - informational supplement (M100-S230) 23. Pennsylvania: Wayne; 2013. [Google Scholar]
- 24.EUCAST (European Committee on Antimicrobial Susceptibility Testing) (2013) Guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance, version 1.0 Sweden.
- 25.Belaaouaj A, Lapoumeroulie C, Caniça MM, Vedel G, Névot P, Krishnamoorthy R, Paul G. Nucleotide sequences of the genes coding for the tem-like beta-lactamases IRT-1 and IRT-2 (formerly called Tri-1 and Tri-2) FEMS Microbiol Lett. 1994;120:75–80. doi: 10.1111/j.1574-6968.1994.tb07010.x. [DOI] [PubMed] [Google Scholar]
- 26.M’zali FM, Gascoyne-Binzi DM, Heritage J, Hawkey PM. Brief reports Detection of mutations conferring extended-spectrum activity on SHV/beta-lactamases using polymerase chain reaction single strand conformational polymorphism ( PCR-SSCP) J Antimicrob Chemother. 1996;37:797–802. doi: 10.1093/jac/37.4.797. [DOI] [PubMed] [Google Scholar]
- 27.Féria C, Ferreira E, Correia JD, Gonçalves J, Caniça M. Patterns and mechanisms of resistance to beta-lactams and beta-lactamase inhibitors in uropathogenic Escherichia coli isolated from dogs in Portugal. J Antimicrob Chemother. 2002;49:77–85. doi: 10.1093/jac/49.1.77. [DOI] [PubMed] [Google Scholar]
- 28.Edelstein M, Pimkin M, Palagin I, Edelstein I, Stratchounski L. Prevalence and molecular epidemiology of CTX-M extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob Agents Chemother. 2003;47:3724–3732. doi: 10.1128/AAC.47.12.3724-3732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 30.Santos DMS, Berchieri A, Fernandes SA, Tavechio AT, Do Amaral LA. Salmonella em carcaças de frango congeladas. Pesqui Vet Bras. 2000;20:39–42. doi: 10.1590/S0100-736X2000000100005. [DOI] [Google Scholar]
- 31.Ribeiro AR, Kellermann A, Dos Santos LR, Bessa MC, Nascimento VP. Salmonella spp. in raw broiler parts: Occurrence, antimicrobial resistance profile and phage typing of the Salmonella Enteritidis isolates. Braz J Microbiol. 2007;38:296–299. doi: 10.1590/S1517-83822007000200021. [DOI] [Google Scholar]
- 32.Yamatogi RS, Oliveira HC, Possebon FS, Pantoja JCF, Joaquim JGF, Pinto JPAN, Araújo JP. Qualitative and quantitative determination and resistance patterns of Salmonella from poultry carcasses. J Food Prot. 2016;79:950–955. doi: 10.4315/0362-028X.JFP-15-489. [DOI] [PubMed] [Google Scholar]
- 33.USDA/FSIS (2012) The nationwide microbiological baseline data collection program: raw chicken parts survey (RCPBS), January 2012 – August 2012. https://www.fsis.usda.gov/shared/PDF/Baseline_Data_Raw_Chicken_Parts.pdf. Accessed 10 December 2016.
- 34.Gonçalves-Tenório A, Silva BN, Rodrigues V, Cadavez V, Gonzales-Barron U. Prevalence of pathogens in poultry meat: a meta-analysis of European published surveys. Foods. 2018;7:2–16. doi: 10.3390/foods7050069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smadi H, Sargeant JM, Shannon HS, Raina P. Growth and inactivation of Salmonella at low refrigerated storage temperatures and thermal inactivation on raw chicken meat and laboratory media: mixed effect meta-analysis. J Epidemiol Glob Health. 2012;2:165–179. doi: 10.1016/j.jegh.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Juneja VK, Eblen BS, Ransom GM. Thermal inactivation of Salmonella spp. in chicken broth, beef, pork, turkey, and chicken: determination of D- and Z values. J Food Sci. 2001;66:146–152. doi: 10.1111/j.1365-2621.2001.tb15597.x. [DOI] [Google Scholar]
- 37.Motta SPO, Flint S, Perry P, Noble A. Consumer contribution to food contamination in Brazil: modelling the food safety risk in the home. Braz J Food Technol. 2014;17:154–165. doi: 10.1590/bjft.2014.018. [DOI] [Google Scholar]
- 38.Soares VM, Pereira JG, Viana C, Izidoro TB, Bersot LS, Pinto JPAN. Transfer of Salmonella Enteritidis to four types of surfaces after cleaning procedures and cross-contamination to tomatoes. Food Microbiol. 2012;30:453–456. doi: 10.1016/j.fm.2011.12.028. [DOI] [PubMed] [Google Scholar]
- 39.Mion L, Parizotto L, Santos LA, Webber B, Cisco IC, Pilotto F, Rodrigues LB, Nascimento VP, Santos LR. Salmonella spp. Isolated by miniaturized most probable number and conventional microbiology in poultry slaughterhouses. Acta Sci Vet. 2016;44:1–5. [Google Scholar]
- 40.Santos LA, Mion L, Marotzki M, Parizotto L, Rodrigues LB, Nascimento VP, Santos LR. Número mais provável miniaturizado e microbiologia convencional para isolamento de Salmonella spp. em abatedouros de frangos de corte. Pesqui Vet Bras. 2015;35:223–229. doi: 10.1590/S0100-736X2015000300003. [DOI] [Google Scholar]
- 41.Lee S, Choi D, Kim H, Kim D, Seo K. Prevalence, seasonal occurrence, and antimicrobial resistance of Salmonella spp. Isolates recovered from chicken carcasses sampled at major poultry processing plants of South Korea Foodborne. Pathog Dis. 2016;13:544–550. doi: 10.1089/fpd.2016.2144. [DOI] [PubMed] [Google Scholar]
- 42.Oscar TP, Rutto GK, Ludwig JB, Parveen S. Qualitative map of Salmonella contamination on young chicken carcasses. J Food Prot. 2010;73:1596–1603. doi: 10.4315/0362-028X-73.9.1596. [DOI] [PubMed] [Google Scholar]
- 43.Brazil (2017) Ministry of Agriculture, Livestock and Supply. Normative Instruction n 08, of 17 February 2017. http://www.imprensanacional.gov.br/materia/-/asset_publisher/Kujrw0TZC2Mb/content/id/20472445/do1-2017-03-03-instrucao-normativa-n-8-de-17-de-fevereiro-de-2017-20472317. Accessed 23 September 2017.
- 44.Brazil (2003) Ministry of Agriculture, Livestock and Supply. Normative Instruction n 78, of 3 November 2003. http://www.adepara.pa.gov.br/sites/default/files/INSTRU% C3%87%C3%83O%20NORMATIVA%20N%C2%BA%2078%2C%20SALMONELLA_0.pdf. Accessed 10 August 2015.
- 45.Kanashiro AMI, Stoppa GFZ, Cardoso ALSP, Tessari ENC, Castro AGM. Serovars of Salmonella spp. isolated from broiler chickens and commercial breeders in diverse regions in Brazil from July 1997 to December 2004. Rev Bras Cienc Avic. 2005;7:195–198. doi: 10.1590/S1516-635X2005000300010. [DOI] [Google Scholar]
- 46.Leal NC, Sá AT, Solari CA, Silva SJ, Hofer E. Sorovares de Salmonella isolados de processos entéricos humanos em Recife-Pernambuco, durante o triênio 1978-1980. Mem Inst Oswaldo Cruz. 1987;82:43–49. doi: 10.1590/S0074-02761987000100007. [DOI] [PubMed] [Google Scholar]
- 47.Hofer E, Zamora MRN, Lopes AE, Moura AMC, Araújo HL, Leite JDD, Leite MDD, Silva Filho SJ. Sorovares de Salmonella em carne de equídeos abatidos no nordeste do Brasil. Pesqui Vet Bras. 2000;20:80–84. doi: 10.1590/S0100-736X2000000200005. [DOI] [Google Scholar]
- 48.McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, Ayers SL, Lam C, Tate HP, Zhao S. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother. 2016;60:5515–5520. doi: 10.1128/AAC.01030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pribul BR, Festivo ML, Rodrigues MS, Costa RG, dos Rodrigues ECP, de Souza MMS, dos Rodrigues DP. Characteristics of quinolone resistance in Salmonella spp. isolates from the food chain in Brazil. Front Microbiol. 2017;8:299. doi: 10.3389/fmicb.2017.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ribeiro VB, Lincopan N, Landgraf M, Franco BDGM, Destro MT. Characterization of class 1 integrons and antibiotic resistance genes in multidrug-resistant Salmonella enterica isolates from foodstuff and related sources. Braz J Microbiol. 2011;42:685–692. doi: 10.1590/S1517-838220110002000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang B, Qiao L, Zhang X, Cui Y, Xia X, Cui S, Wang X, Meng X, Ge W, Shi X, Wang D, Meng J. Serotyping, antimicrobial susceptibility, pulsed field gel electrophoresis analysis of Salmonella isolates from retail foods in Henan Province. China Food Control. 2013;32:228–235. doi: 10.1016/j.foodcont.2012.11.022. [DOI] [Google Scholar]
- 52.Stevenson JE, Gay K, Barrett TJ, Medalla F, Chiller TM, Angulo FJ. Increase in nalidixic acid resistance among non-Typhi Salmonella enterica isolates in the United States from 1996 to 2003. Antimicrob Agents Chemother. 2003;51:195–195. doi: 10.1128/AAC.00222-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Casin I, Breuil J, Darchis JP, Guelpa C, Collatz E. Fluoroquinolone resistance linked to GyrA, GyrB, and ParC mutations in Salmonella enterica typhimurium isolates in humans. Emerg Infect Dis. 2003;9:1455–1457. doi: 10.3201/eid0911.030317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eaves DJ, Randall L, Gray DT, Buckley A, Woodward MJ, White AP, Piddock LJV. Prevalence of mutations within the quinolone resistance-determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone-resistant Salmonella enterica. Antimicrob Agents Chemother. 2004;48:4012–4015. doi: 10.1128/AAC.48.10.4012-4015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parry CM, Threlfall EJ. Antimicrobial resistance in typhoidal and nontyphoidal salmonellae. Curr Opin Infect Dis. 2008;21:531–538. doi: 10.1097/QCO.0b013e32830f453a. [DOI] [PubMed] [Google Scholar]
- 56.Pribul BR, Festivo ML, Rodrigues MS, Costa RG, Rodrigues ECP, de Souza MMS, Rodrigues DP (2017) Characteristics of quinolone resistance in Salmonella spp. Isolates from the food chain in Brazil. Front. Microbiol. 8:299. 10.3389/fmicb.2017.00299 [DOI] [PMC free article] [PubMed]
- 57.Almeida F, Seribelli AA, Medeiros MIC, DDP R, de Mello Varani AD, Luo Y, Allard MW, Falcão JP. Phylogenetic and antimicrobial resistance gene analysis of Salmonella Typhimurium strains isolated in Brazil by whole genome sequencing. PLoS One. 2018;13:1–16. doi: 10.1371/journal.pone.0201882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fitch FM, Carmo-Rodrigues MS, Oliveira VG, Gaspari MV, Dos Santos A, De Freitas JB, Pignatari AC. β-Lactam resistance genes: characterization, epidemiology, and first detection of blaCTX-M-1 and blaCTX-M-14 in Salmonella spp. Isolated from poultry in Brazil-Brazil Ministry of Agriculture’s Pathogen Reduction Program. Microb. Drug Resist. 2016;22:164–171. doi: 10.1089/mdr.2015.0143. [DOI] [PubMed] [Google Scholar]
- 59.Fonzé E, Charlier P, To’th Y, Vermeire M, Raquet X, Dubus A, Frère JM. TEM1 Beta-lactamase structure solved by molecular replacement and refined structure of the S235A mutant. Acta Crystallogr D Biol Crystallogr. 1995;51:682–694. doi: 10.1107/S0907444994014496. [DOI] [PubMed] [Google Scholar]
- 60.Falagas ME, Karageorgopoulos DE. Extended-spectrum beta-lactamase-producing organisms. J Hosp Infect. 2009;73:345–354. doi: 10.1016/j.jhin.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 61.Peirano G, Agerso Y, Aarestrup FM, Dos Reis EM, Rodrigues DP. Occurrence of integrons and antimicrobial resistance genes among Salmonella enterica from Brazil. J Antimicrob Chemother. 2006;58:305–309. doi: 10.1093/jac/dkl248. [DOI] [PubMed] [Google Scholar]
- 62.Ziech RE, Lampugnani C, Perin AP, Sereno MJ, Sfaciotte RAP, Viana C, Soares VM, Pinto JPAN, Bersot LS. Multidrug resistance and ESBL-producing Salmonella spp. isolated from broiler processing plants. Braz J Microbiol. 2016;47:191–195. doi: 10.1016/j.bjm.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brazil (1998) Ministry of Agriculture, Livestock and Supply. Ordinance n 448, of 10 September 1998. http://www.sgc.goias.gov.br/upload/arquivos/2012-01/pamvet.pdf. Accessed 11 March 2016.
- 64.Scur MC, Pinto FGS, Bona EAM, Weber LD, Alves LFA, Moura AC. Occurrence and antimicrobial resistance of Salmonella serotypes isolates recovered from poultry of Western Paraná. Brazil Afr J Agric Res. 2014;9:823–830. doi: 10.5897/AJAR2013.8202. [DOI] [Google Scholar]
- 65.Cortez ALL, Carvalho ACFB, Ikuno AA, Bürger KP, Vidal-Martins AMC. Resistência antimicrobiana de cepas de Salmonella spp. isoladas de abatedouros de aves. Arq Inst Biol. 2006;73:157–163. [Google Scholar]
- 66.Tennant SM, Maclennan CA, Simon R, Martin LB, Khan MI. Nontyphoidal Salmonella disease: current status of vaccine research and development. Vaccine. 2016;34:2907–2910. doi: 10.1016/j.vaccine.2016.03.072. [DOI] [PubMed] [Google Scholar]