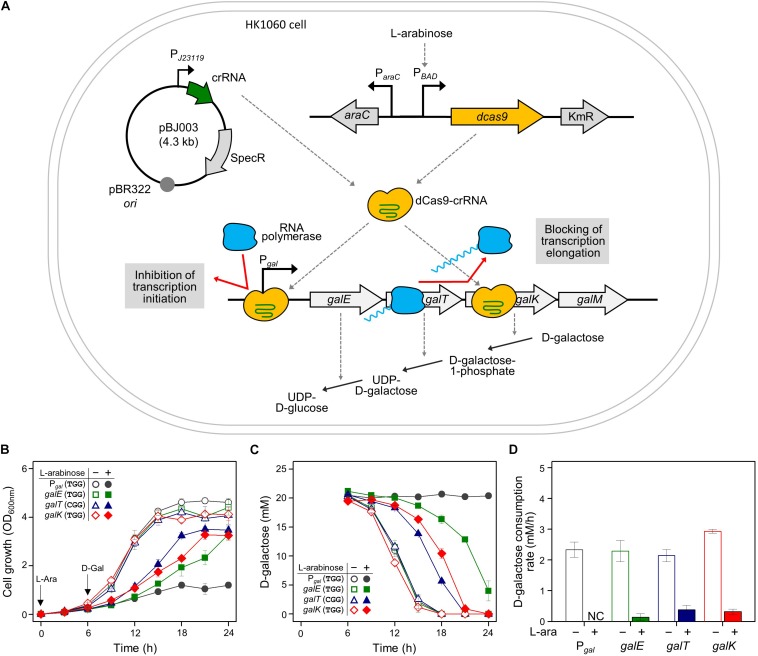
FIGURE 1.
Regulation of gal operon and control of D-galactose metabolic rates by CRISPRi. (A) Expression of gal promoter and structural genes regulated by dCas9-crRNAs. The control of the gal promoter and structural galETK genes was used as a model system for comparing the transcriptional initiation and elongation by CRISPRi. The target DNAs in the gal operon can be attached by active dCas9-crRNA complexes, which are formed by dCas9 proteins expressed in the presence of non-metabolizable L-arabinose, and crRNAs transcribed constitutively in cells. D-galactose metabolism depends on the expression of D-galactose metabolic enzymes in the gal operon. (B) Growth profiles of HK1060 cells carrying specific crRNAs targeting gal promoter and structural galETK genes. Retarded cell growth was observed due to the repression of galactose enzymes by L-arabinose. (C) Change of residual D-galactose concentration in culture media. D-galactose was added at 6 h and monitored until the late stationary growth phase. (D) D-galactose consumption rates affected by CRISPRi. The concentration changes of residual D-galactose between 9 and 12 h were used for the calculation of D-galactose consumption rates. Arrows indicate when L-arabinose (if needed) and D-galactose were added. NC, not consumed.