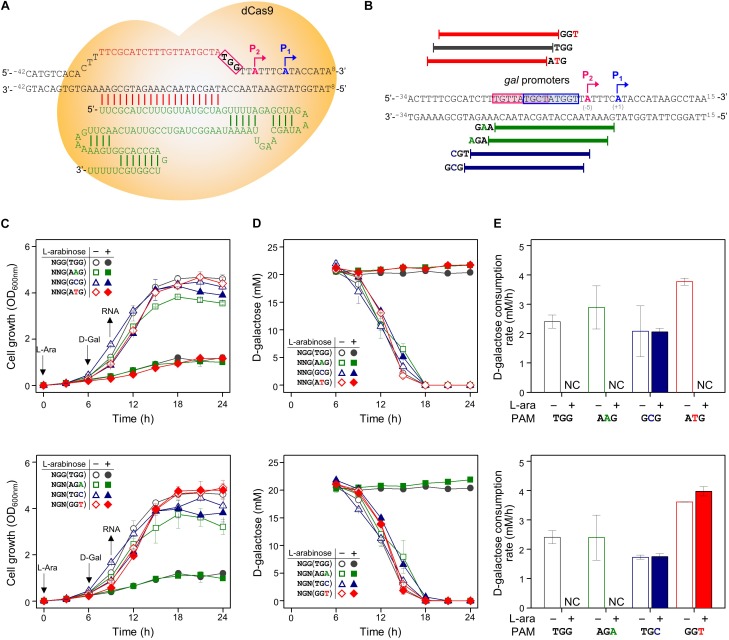
FIGURE 2.
Tight repression of the gal promoter by CRISPRi with modified PAM sequences. (A) The inability of the RNA polymerase to recognize and attach to the gal promoter region, which is occupied by the dCas9-crRNA complex, inhibits the initiation of transcription. Blue and red boxes indicate overlapped –10 regions of P1 and P2 promoters, respectively. The gal promoter-specific crRNA was transcribed from plasmid pBJ005. (B) Design of target DNA sequences with modified PAM sequences in gal promoter. The gal P1 and P2 promoters overlapped each other. The modified PAM sequences of NNG (top) and NGN (bottom) were evaluated by growth profiles (C), utilization of D-galactose (D), and comparison of D-galactose consumption rates (E). D-galactose was added at 6 h, and the concentration changes of residual D-galactose between 9 and 12 h were used to calculate D-galactose consumption rates. NC, not consumed. Arrows indicate when L-arabinose (if needed) and D-galactose were added, and when samples were taken for RNA extraction.