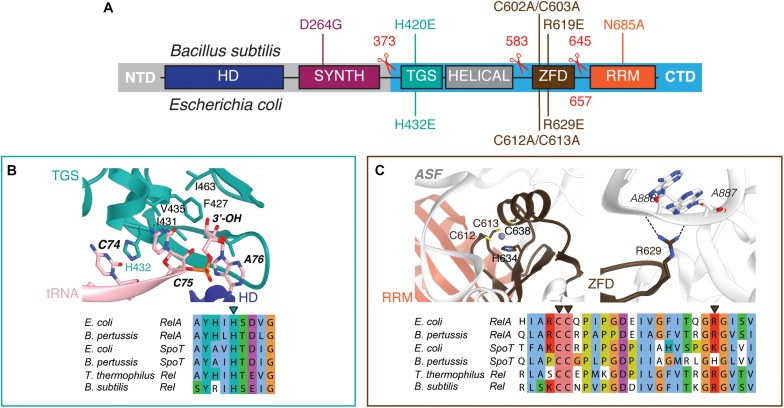
FIGURE 1.
Domain structure of ‘long’ ribosome-associated RSHs Rel and RelA. (A) The NTD region contains (p)ppGpp hydrolysis (HD) and (p)ppGpp synthesis (SYNTH) NTD domains. TGS (ThrRS, GTPase and SpoT), Helical, ZFD (Zinc Finger Domain) and RRM (RNA Recognition Motif) domains comprise the regulatory CTD region. Mutations and truncations of B. subtilis Rel and E. coli RelA used in this study are indicated above and below the domain schematics, respectively. (B) Conservation and structural environment of mutations in the TGS domain used in the current study. (C) Conservation and structural environment of mutations in the RRM domain used in the current study. The 3D structures are as per from Loveland and colleagues (Loveland et al., 2016), RDB accession number 5KPX.