FIGURE 4.
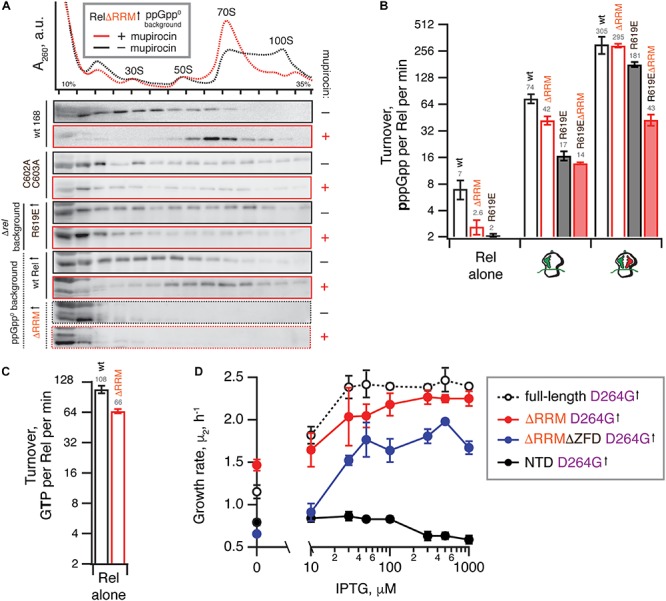
Deletion of the regulatory RRM domain destabilizes Rel binding to starved ribosomal complexes and does not abrogate the hydrolysis activity. (A) Polysome profile and immunoblot analyses of Rel variants expressed either ectopically (↑) under the control of IPTG-inducible Phy–spnak promotor [ΔRRM Rel in ppGpp0 B. subtilis (VHB184), R619E Rel in Δrel B. subtilis (VHB282)] or from the native chromosomal locus [C602A C603A mutant (VHB144)]. Expression of RelΔRRM was induced by 1 mM IPTG for 10 min followed by a 10 min challenge with 700 nM mupirocin. To drive the expression of R619E Rel, the strain was grown in LB supplemented with 1 mM IPTG. In the case of R619E and C602A C603A Rel the culture was treated with mupirocin for 20 min. Polysome profiles of all tested Rel variants are presented in Supplementary Figure S5, and an uncut version of a representative anti-Rel immunoblot is shown in Supplementary Figure S2G. (B,C) The effect of the RRM deletion on synthetic (B) and hydrolytic activity (C). The effects of the RRM deletion and the R619E mutation on Rel synthetic activity were assayed either alone or in the presence of either initiation and starved ribosomal complexes. The error bars represent standard deviations of the turnover estimates by linear regression using four data points. (D) The effects of titratable expression of synthesis-inactive D264G mutants (full-length VHB156, ΔRRM VHB162, ΔRRMΔZFD VHB163 and RelNTD VHB164) on Δrel B. subtilis growing on liquid LB medium at 37°C. The growth rates (μ2) were calculated from three independent biological replicates and the error bars represent standard deviations.
