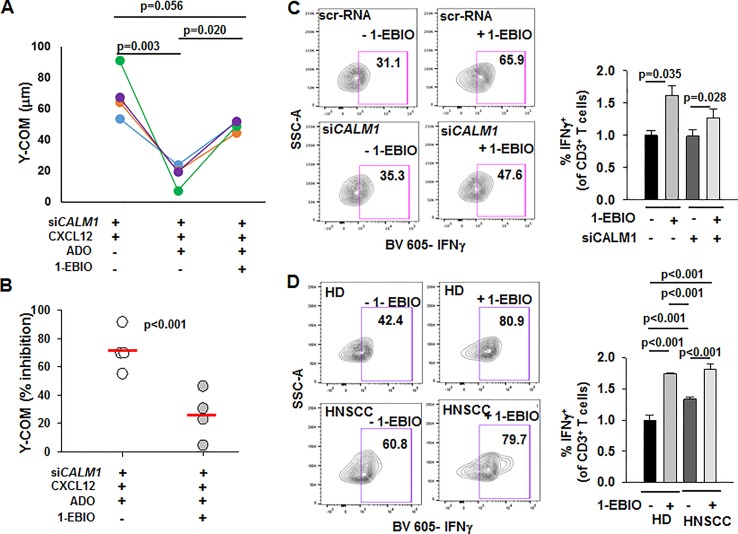
Figure 7.
Potentiation of chemotaxis in the presence of adenosine and INFγ production by 1-EBIO in CALM1-knockdown HD T cells. (A) Y-COM values calculated for HD T cells transfected with siCALM1 migrating along either a CXCL12 gradient or a combination gradient of CXCL12 with adenosine (ADO) with or without preincubation with 20 μM 1-EBIO (n = 4 donors). Transfected cells, identified by positive GFP staining, were tracked for analyzing Y-COM. (B) Percentage inhibition by ADO of Y-COM (B) in cells pretreated with 1-EBIO. Horizontal red line represents mean values for each group. (C) (Left) Representative flow cytometry plots gated on CD3+ T cells transfected with either scr-RNA or siCALM1 with or without 1-EBIO (100 μM) showing IFNγ production after stimulation. Transfected cells were identified by positive staining for GFP. (Right) Quantification of IFNγ induction in scr-RNA or siCALM1 transfected CD3+ T cells with or without 1-EBIO in three HD. The data were normalized to IFNγ production in control scr-RNA transfected HD cells. Bars represent mean ± SEM. (D) (Left) Representative flow cytometry plots gated on HD or HNSCC CD3+ T cells with or without 1-EBIO (100 μM) showing IFNγ production after stimulation. (Right) Quantification of IFNγ-producing HD and HNSCC T cells after stimulation in the presence or absence of 1-EBIO. The data were normalized to IFNγ production in control HD cells. For the HD + 1-EBIO condition, bars represent mean ± range for two individuals, while the rest of the bars in the graph represent mean ± SD for data from three HD and HNSCC patients. For analysis of (C, D), live cell population was identified by exclusion from Zombie Aqua live/dead stain and gated on CD3+ population and transfected cells were identified by positive staining for GFP (in panel C). IFNγ+ T cell populations were selected by drawing rectangle gates. The numbers in the gates are the percentages of IFNγ+ CD3+ T cells. Data in (A) were analyzed with one-way repeated measures ANOVA (p <0.001), in (B) with paired Student's t test, and in (C) with a two-way ANOVA [p = 0.207 for scr-RNA vs. siCALM1 and p = 0.006 for – 1-EBIO vs. +1-EBIO]. Data in (D) were analyzed by Kruksal-Wallis One Way ANOVA on Ranks (p < 0.001).