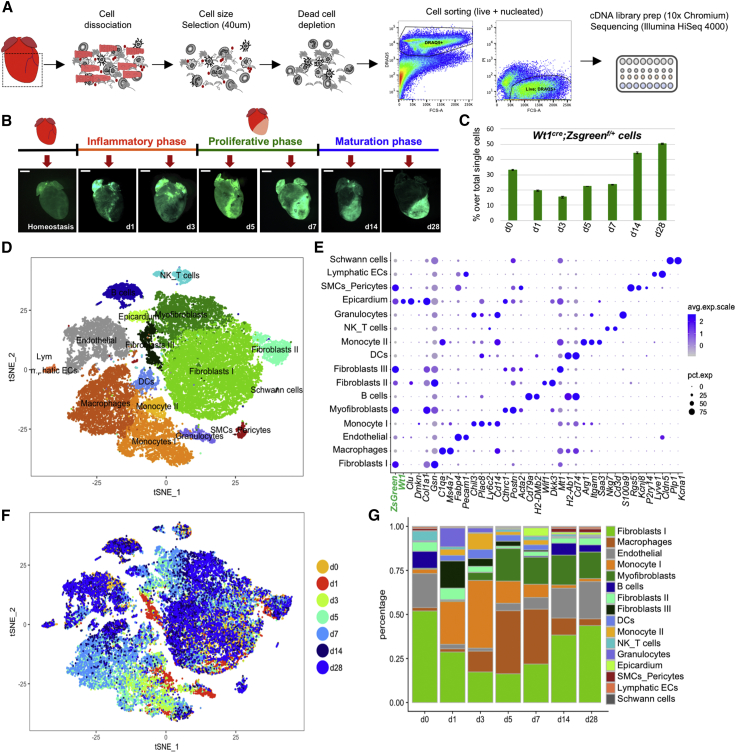
Figure 1.
Single-Cell RNA-Seq of Interstitial Cells Post-MI Reveals Population Dynamics in Cardiac Repair
(A) Murine cardiac interstitial cells were isolated by mechanical and enzymatic dissociation of adult mouse cardiac ventricular tissue (dashed square); mesh purification, magnetic dead cell depletion, and sorting were performed to exclude cardiomyocytes and apoptotic or necrotic cells before analysis. A total of 38,600 cells were captured and sequenced (n = 7 mice).
(B) Selected time points and whole-mount images of Wt1cre;ZsGreenf/+ mice used to trace epicardial-derived components in the cardiac interstitium.
(C) Percentage of single live nucleated ZsGreen+ interstitial cells detected by flow cytometry in the samples used for scRNAseq. Data shown as mean ± SD of two technical replicates at each time point.
(D) t-Distributed stochastic neighbor embedding (t-SNE) plot of the aggregate of all sequenced cells across time points. Seurat analysis with 24 PC and resolution 0.5 was used to define 16 main clusters.
(E) Dot-plot visualization of top marker genes used to identify clusters. Dot sizes denote percentage of expression per cluster; color gradient defines average expression per cell.
(F) t-SNE plot showing cell contribution by time point identified by color.
(G) Bar plot of percentage of cluster contributions per time point.
See also Figures S1 and S2 and Tables S1 and S2.