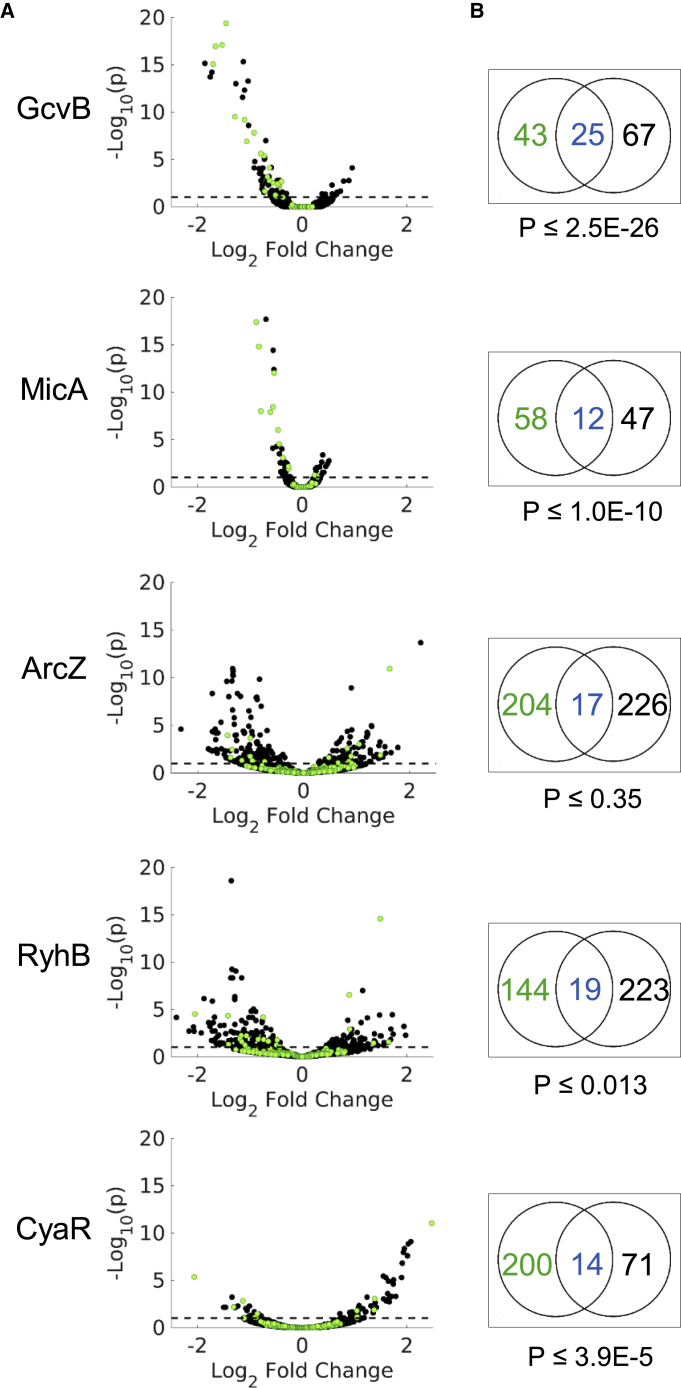
Figure 2.
Measuring the Effect of the sRNA on Target mRNA Levels
(A) Only a subset of the targets shows an expression change following sRNA overexpression. Shown are Volcano plots of RNA-seq results of gene expression change following sRNA overexpression. Gene expression change is represented by the log2 fold change in expression levels, as obtained from DESeq2 analysis (Love et al., 2014) (x axis). The statistical significance of the change is represented as −log10p (y axis). p is the p value corrected for multiple hypothesis testing (padj from DESeq2). For clarity, only genes with −log10p ≤ 20 are presented. Green dots represent the sRNA targets that were detected by RIL-seq applied to E. coli grown to a certain growth phase or condition (GcvB to exponential phase; MicA, ArcZ, and CyaR to stationary phase; and RyhB to exponential phase under iron limitation). Black dots represent the rest of the E. coli genes. The dashed line represents the statistical significance threshold (p ≤ 0.1).
(B) sRNA targets detected by RIL-seq were enriched among genes showing a statistically significant change in expression level following overexpression of the sRNA (statistical significance of the enrichment was computed by hypergeometric test). Black numbers represent non-target genes showing a statistically significant change in expression (both up- and downregulated). Blue/green numbers represent RIL-seq targets that showed/did not show a statistically significant change in expression.
See also Figures S1, S3, and S4, and Tables S1, S2, S3, and S4.