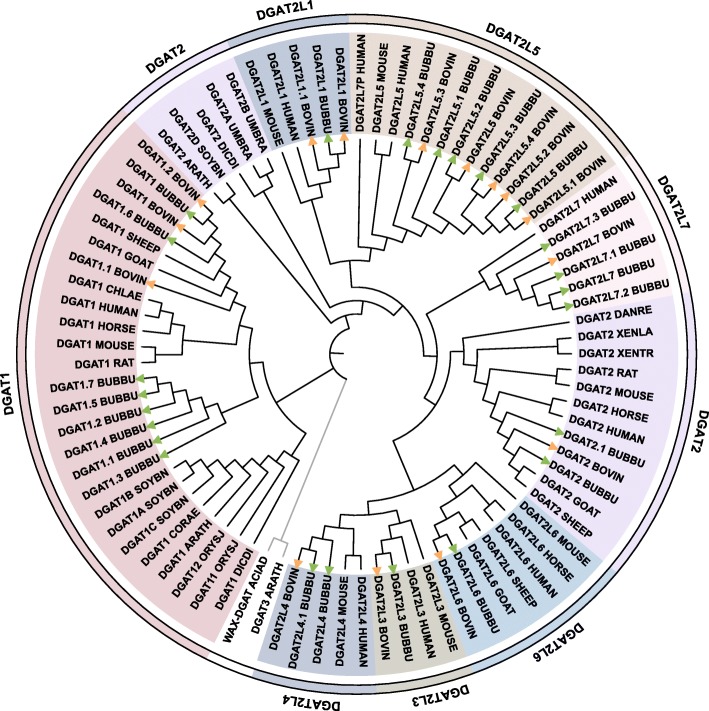
Fig. 2.
Phylogenetic Neighbor-Joining (NJ) tree of DGAT proteins from different organisms. Identified DGATs in Bubalus bubalis (BUBBU) and Bos taurus (BOVIN) together with verified DGATs from Capra hircus (GOAT), Ovis aries (SHEEP), Equus caballus (HORSE), Homo sapiens (HUMAN), Mus musculus (MOUSE), Rattus norvegicus (RAT), Arabidopsis thaliana (ARATH), Chlorocebus aethiops (CHLAE), Dictyostelium discoideum (DICDI), Corylus americana (CORAE), Oryza sativa subsp. japonica (ORYSJ), Glycine max (SOYBN), Danio rerio (DANRE), Xenopus laevis (XENLA), Xenopus tropicalis (XENTR), Umbelopsis ramanniana (UMBRA) and Acinetobacter baylyi (ACIAD) were included in the analyses. The DGAT enzymes are grouped into eight clusters including DGAT1, DGAT2 and DGAT2L1, DGAT2L3, DGAT2L4, DGAT2L5, DGAT2L6 and DGAT2L7, which are represented by different colors. The green and orange arrows represent the identified DGAT proteins isoforms from buffalo and cattle